Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Miled, N., Yan, Y., Hon, W.C., Perisic, O., Zvelebil, M., Inbar, Y., Schneidman-Duhovny, D., Wolfson, H.J., Backer, J.M., Williams, R.L.(2007) Science 317: 239
- PubMed: 17626883
- DOI: https://doi.org/10.1126/science.1135394
- Primary Citation of Related Structures:
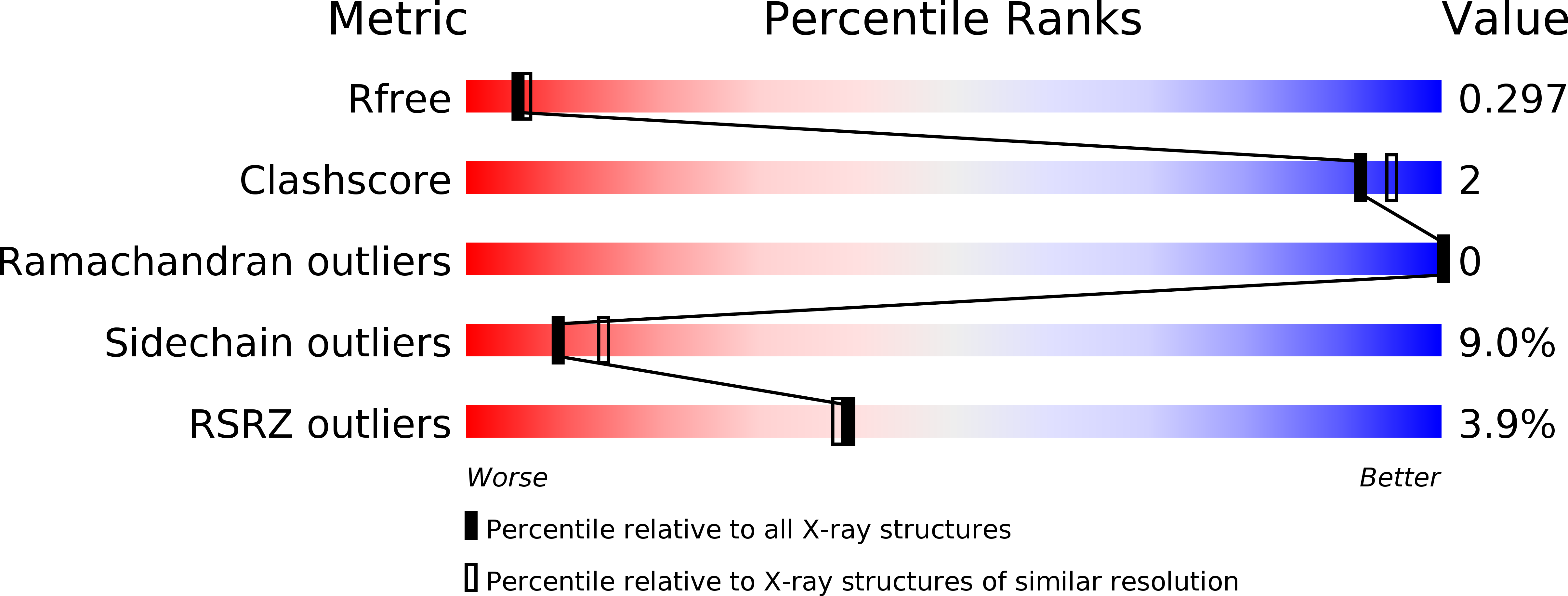
2V1Y - PubMed Abstract:
Many human cancers involve up-regulation of the phosphoinositide 3-kinase PI3Kalpha, with oncogenic mutations identified in both the p110alpha catalytic and the p85alpha regulatory subunits. We used crystallographic and biochemical approaches to gain insight into activating mutations in two noncatalytic p110alpha domains-the adaptor-binding and the helical domains. A structure of the adaptor-binding domain of p110alpha in a complex with the p85alpha inter-Src homology 2 (inter-SH2) domain shows that oncogenic mutations in the adaptor-binding domain are not at the inter-SH2 interface but in a polar surface patch that is a plausible docking site for other domains in the holo p110/p85 complex. We also examined helical domain mutations and found that the Glu545 to Lys545 (E545K) oncogenic mutant disrupts an inhibitory charge-charge interaction with the p85 N-terminal SH2 domain. These studies extend our understanding of the architecture of PI3Ks and provide insight into how two classes of mutations that cause a gain in function can lead to cancer.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.