Nuclear Import Mechanism of the Ejc Component Mago- Y14 Revealed by Structural Studies of Importin 13.
Bono, F., Cook, A.G., Grunwald, M., Ebert, J., Conti, E.(2010) Mol Cell 37: 211
- PubMed: 20122403
- DOI: https://doi.org/10.1016/j.molcel.2010.01.007
- Primary Citation of Related Structures:
2X19, 2X1G - PubMed Abstract:
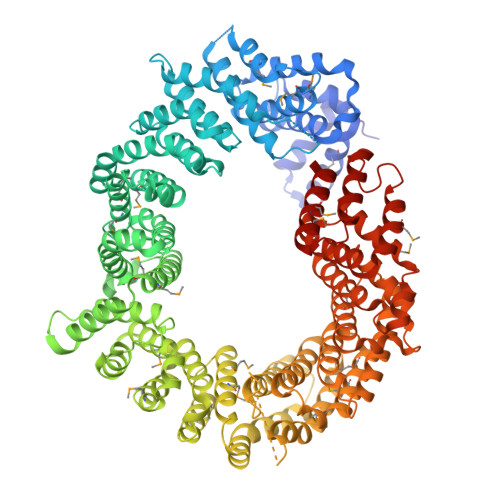
Mago and Y14 are core components of the exon junction complex (EJC), an assembly central to nonsense-mediated mRNA decay in humans and mRNA localization in flies. The Mago-Y14 heterodimer shuttles between the nucleus, where it is loaded onto specific mRNAs, and the cytoplasm, where it functions in translational regulation. The heterodimer is imported back into the nucleus by Importin 13 (Imp13), a member of the karyopherin-beta family of transport factors. We have elucidated the structural basis of the Mago-Y14 nuclear import cycle. The 3.35 A structure of the Drosophila Imp13-Mago-Y14 complex shows that Imp13 forms a ring-like molecule, reminiscent of Crm1, and encircles the Mago-Y14 cargo with a conserved interaction surface. The 2.8 A structure of human Imp13 bound to RanGTP reveals how Mago-Y14 is released in the nucleus by a steric hindrance mechanism. Comparison of the two structures suggests how this unusual karyopherin might function in bidirectional nucleocytoplasmic transport.
- Max-Planck-Institute of Biochemistry, Department of Structural Cell Biology, Am Klopferspitz 18, 82152 Martinsried, Germany.
Organizational Affiliation: