A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
Montanier, C.Y., Correia, M.A.S., Flint, J.E., Zhu, Y., Basle, A., Mckee, L.S., Prates, J.A.M., Polizzi, S.J., Coutinho, P.M., Lewis, R.J., Henrissat, B., Fontes, C.M.G.A., Gilbert, H.J.(2011) J Biol Chem 286: 22499
- PubMed: 21454512
- DOI: https://doi.org/10.1074/jbc.M110.217372
- Primary Citation of Related Structures:
2YB7, 2YFU, 2YFZ, 2YG0 - PubMed Abstract:
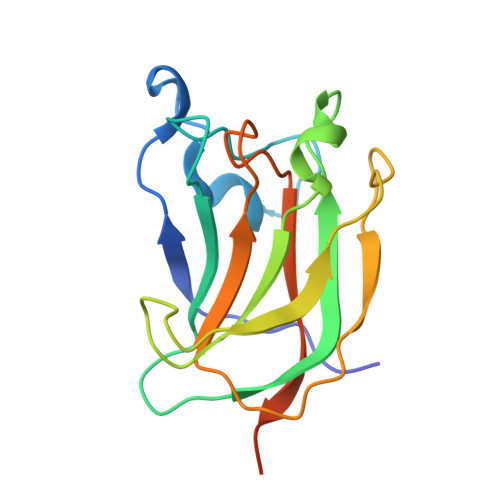
The enzymic degradation of plant cell walls plays a central role in the carbon cycle and is of increasing environmental and industrial significance. The catalytic modules of enzymes that catalyze this process are generally appended to noncatalytic carbohydrate-binding modules (CBMs). CBMs potentiate the rate of catalysis by bringing their cognate enzymes into intimate contact with the target substrate. A powerful plant cell wall-degrading system is the Clostridium thermocellum multienzyme complex, termed the "cellulosome." Here, we identify a novel CBM (CtCBM62) within the large C. thermocellum cellulosomal protein Cthe_2193 (defined as CtXyl5A), which establishes a new CBM family. Phylogenetic analysis of CBM62 members indicates that a circular permutation occurred within the family. CtCBM62 binds to d-galactose and l-arabinopyranose in either anomeric configuration. The crystal structures of CtCBM62, in complex with oligosaccharides containing α- and β-galactose residues, show that the ligand-binding site in the β-sandwich protein is located in the loops that connect the two β-sheets. Specificity is conferred through numerous interactions with the axial O4 of the target sugars, a feature that distinguishes galactose and arabinose from the other major sugars located in plant cell walls. CtCBM62 displays tighter affinity for multivalent ligands compared with molecules containing single galactose residues, which is associated with precipitation of these complex carbohydrates. These avidity effects, which confer the targeting of polysaccharides, are mediated by calcium-dependent oligomerization of the CBM.
Organizational Affiliation:
Centro de Investigação Interdisciplinar em Sanidade Animal, Faculdade de Medicina Veterinária, Universidade Técnica de Lisboa, Avenida da Universidade Técnica, 1300-477 Lisboa, Portugal.