Characterization of 3-ketosteroid 9{alpha}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis
Capyk, J.K., D'Angelo, I., Strynadka, N.C., Eltis, L.D.(2009) J Biological Chem 284: 9937-9946
- PubMed: 19234303
- DOI: https://doi.org/10.1074/jbc.M900719200
- Primary Citation of Related Structures:
2ZYL - PubMed Abstract:
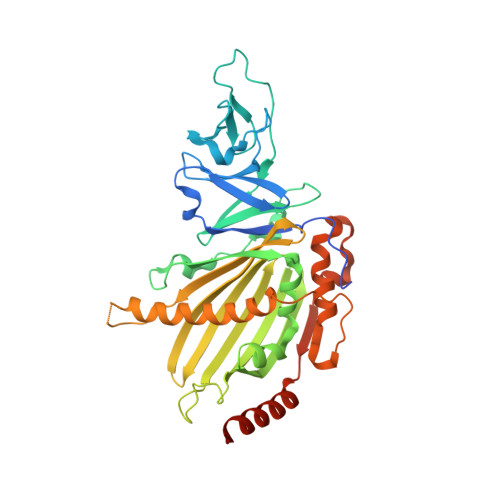
KshAB (3-Ketosteroid 9alpha-hydroxylase) is a two-component Rieske oxygenase (RO) in the cholesterol catabolic pathway of Mycobacterium tuberculosis. Although the enzyme has been implicated in pathogenesis, it has largely been characterized by bioinformatics and molecular genetics. Purified KshB, the reductase component, was a monomeric protein containing a plant-type [2Fe-2S] cluster and FAD. KshA, the oxygenase, was a homotrimer containing a Rieske [2Fe-2S] cluster and mononuclear ferrous iron. Of two potential substrates, reconstituted KshAB had twice the specificity for 1,4-androstadiene-3,17-dione as for 4-androstene-3,17-dione. The transformation of both substrates was well coupled to the consumption of O(2). Nevertheless, the reactivity of KshAB with O(2) was low in the presence of 1,4-androstadiene-3,17-dione, with a k(cat)/K(m)(O(2)) of 2450 +/- 80 m(-1) s(-1). The crystallographic structure of KshA, determined to 2.3A(,) revealed an overall fold and a head-to-tail subunit arrangement typical of ROs. The central fold of the catalytic domain lacks all insertions found in characterized ROs, consistent with a minimal and perhaps archetypical RO catalytic domain. The structure of KshA is further distinguished by a C-terminal helix, which stabilizes subunit interactions in the functional trimer. Finally, the substrate-binding pocket extends farther into KshA than in other ROs, consistent with the large steroid substrate, and the funnel accessing the active site is differently orientated. This study provides a solid basis for further studies of a key steroid-transforming enzyme of biotechnological and medical importance.
- Departments of Biochemistry and Molecular Biology and Microbiology and Immunology, Life Sciences Institute, University of British Columbia, Vancouver V6 1Z3, Canada.
Organizational Affiliation: