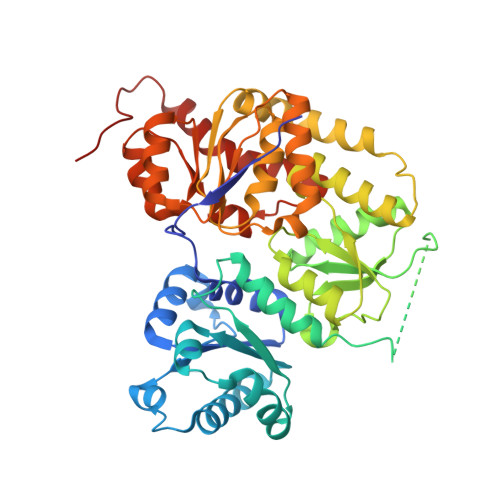
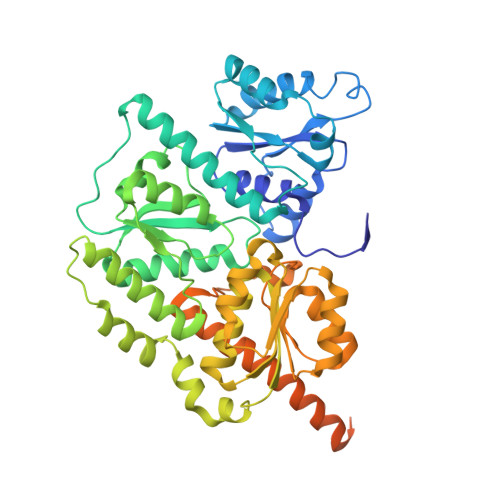
Crystal Structure of the Nitrogenase-Like Dark Operative Protochlorophyllide Oxidoreductase Catalytic Complex (Chln/Chlb)2.
Broecker, M.J., Schomburg, S., Heinz, D.W., Jahn, D., Schubert, W.-D., Moser, J.(2010) J Biol Chem 285: 27336
- PubMed: 20558746
- DOI: https://doi.org/10.1074/jbc.M110.126698
- Primary Citation of Related Structures:
2XDQ - PubMed Abstract:
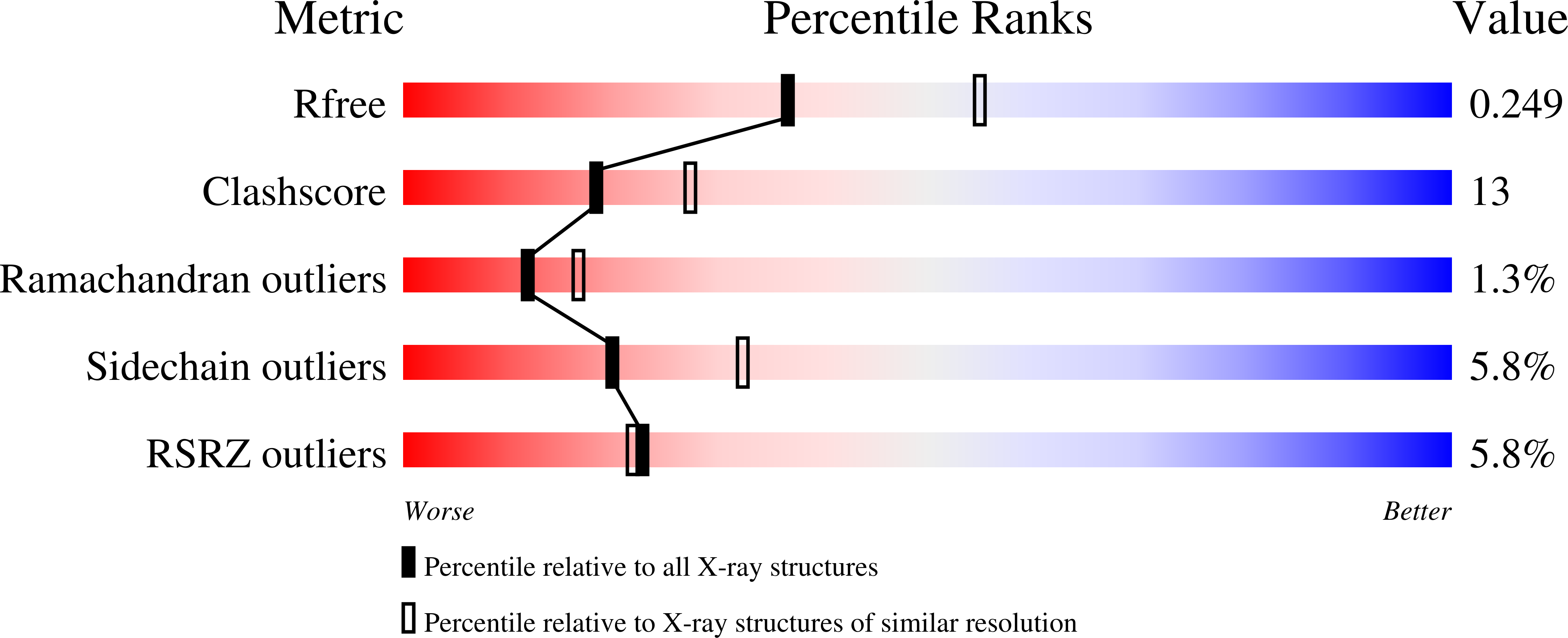
During (bacterio)chlorophyll biosynthesis of many photosynthetically active organisms, dark operative protochlorophyllide oxidoreductase (DPOR) catalyzes the two-electron reduction of ring D of protochlorophyllide to form chlorophyllide. DPOR is composed of the subunits ChlL, ChlN, and ChlB. Homodimeric ChlL(2) bearing an intersubunit [4Fe-4S] cluster is an ATP-dependent reductase transferring single electrons to the heterotetrameric (ChlN/ChlB)(2) complex. The latter contains two intersubunit [4Fe-4S] clusters and two protochlorophyllide binding sites, respectively. Here we present the crystal structure of the catalytic (ChlN/ChlB)(2) complex of DPOR from the cyanobacterium Thermosynechococcus elongatus at a resolution of 2.4 A. Subunits ChlN and ChlB exhibit a related architecture of three subdomains each built around a central, parallel beta-sheet surrounded by alpha-helices. The (ChlN/ChlB)(2) crystal structure reveals a [4Fe-4S] cluster coordinated by an aspartate oxygen alongside three cysteine ligands. Two equivalent substrate binding sites enriched in aromatic residues for protochlorophyllide substrate binding are located at the interface of each ChlN/ChlB half-tetramer. The complete octameric (ChlN/ChlB)(2)(ChlL(2))(2) complex of DPOR was modeled based on the crystal structure and earlier functional studies. The electron transfer pathway via the various redox centers of DPOR to the substrate is proposed.
Organizational Affiliation:
Institut für Mikrobiologie, Technische Universität Braunschweig, Spielmannstrasse 7, D-38106 Braunschweig, Germany.