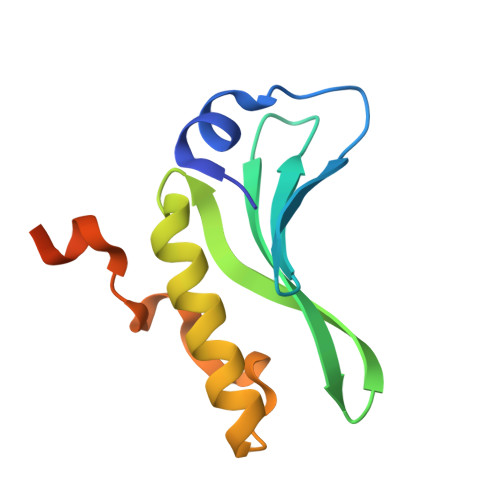
Structures of the arm-type binding domains of HPI and HAI7 integrases
Szwagierczak, A., Antonenka, U., Popowicz, G.M., Sitar, T., Holak, T.A., Rakin, A.(2009) J Biological Chem 284: 31664-31671
- PubMed: 19737930
- DOI: https://doi.org/10.1074/jbc.M109.059261
- Primary Citation of Related Structures:
3JTZ, 3JU0 - PubMed Abstract:
The structures of the N-terminal domains of two integrases of closely related but not identical asn tDNA-associated genomic islands, Yersinia HPI (high pathogenicity island; encoding siderophore yersiniabactin biosynthesis and transport) and an Erwinia carotovora genomic island with yet unknown function, HAI7, have been resolved. Both integrases utilize a novel four-stranded beta-sheet DNA-binding motif, in contrast to the known proteins that bind their DNA targets by means of three-stranded beta-sheets. Moreover, the beta-sheets in Int(HPI) and Int(HAI7) are longer than those in other integrases, and the structured helical N terminus is positioned perpendicularly to the large C-terminal helix. These differences strongly support the proposal that the integrases of the genomic islands make up a distinct evolutionary branch of the site-specific recombinases that utilize a unique DNA-binding mechanism.
- Max Planck Institute for Biochemistry, D-82152 Martinsried, Germany.
Organizational Affiliation: