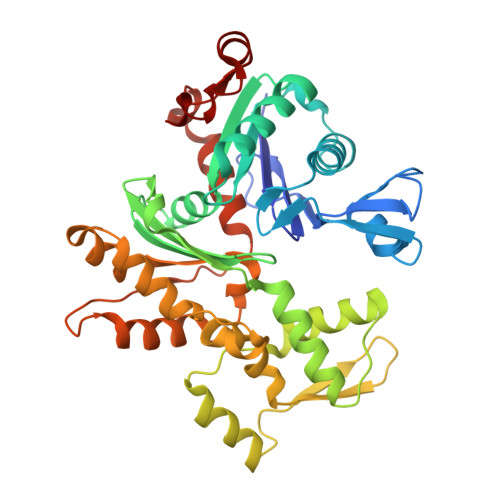
Multiple Forms of Spire-Actin Complexes and their Functional Consequences.
Chen, C.K., Sawaya, M.R., Phillips, M.L., Reisler, E., Quinlan, M.E.(2012) J Biol Chem 287: 10684-10692
- PubMed: 22334675
- DOI: https://doi.org/10.1074/jbc.M111.317792
- Primary Citation of Related Structures:
3UE5, 4EFH - PubMed Abstract:
Spire is a WH2 domain-containing actin nucleator essential for establishing an actin mesh during oogenesis. In vitro, in addition to nucleating filaments, Spire can sever them and sequester actin monomers. Understanding how Spire is capable of these disparate functions and which are physiologically relevant is an important goal. To study severing, we examined the effect of Drosophila Spire on preformed filaments in bulk and single filament assays. We observed rapid depolymerization of actin filaments by Spire, which we conclude is largely due to its sequestration activity and enhanced by its weak severing activity. We also studied the solution and crystal structures of Spire-actin complexes. We find structural and functional differences between constructs containing four WH2 domains (Spir-ABCD) and two WH2 domains (Spir-CD) that may provide insight into the mechanisms of nucleation and sequestration. Intriguingly, we observed lateral interactions between actin monomers associated with Spir-ABCD, suggesting that the structures built by these four tandem WH2 domains are more complex than originally imagined. Finally, we propose that Spire-actin mixtures contain both nuclei and sequestration structures.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California Los Angeles, Los Angeles, California 90095.