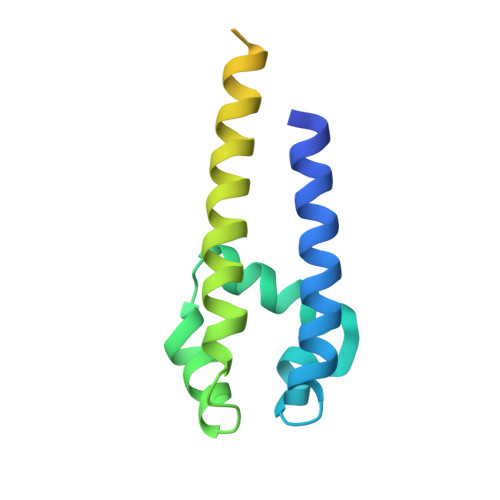
Role of the C-Terminal Domain in the Structure and Function of Tetrameric Sodium Channels.
Bagneris, C., Decaen, P.G., Hall, B.A., Naylor, C.E., Clapham, D.E., Kay, C.W.M., Wallace, B.A.(2013) Nat Commun 4: 2465
- PubMed: 24051986
- DOI: https://doi.org/10.1038/ncomms3465
- Primary Citation of Related Structures:
3ZJZ - PubMed Abstract:
Voltage-gated sodium channels have essential roles in electrical signalling. Prokaryotic sodium channels are tetramers consisting of transmembrane (TM) voltage-sensing and pore domains, and a cytoplasmic carboxy-terminal domain. Previous crystal structures of bacterial sodium channels revealed the nature of their TM domains but not their C-terminal domains (CTDs). Here, using electron paramagnetic resonance (EPR) spectroscopy combined with molecular dynamics, we show that the CTD of the NavMs channel from Magnetococcus marinus includes a flexible region linking the TM domains to a four-helix coiled-coil bundle. A 2.9 Å resolution crystal structure of the NavMs pore indicates the position of the CTD, which is consistent with the EPR-derived structure. Functional analyses demonstrate that the coiled-coil domain couples inactivation with channel opening, and is enabled by negatively charged residues in the linker region. A mechanism for gating is proposed based on the structure, whereby splaying of the bottom of the pore is possible without requiring unravelling of the coiled-coil.
- 1] School of Biological Sciences, Institute of Structural and Molecular Biology, Birkbeck College, University of London, London WC1E 7HX, UK [2].
Organizational Affiliation: