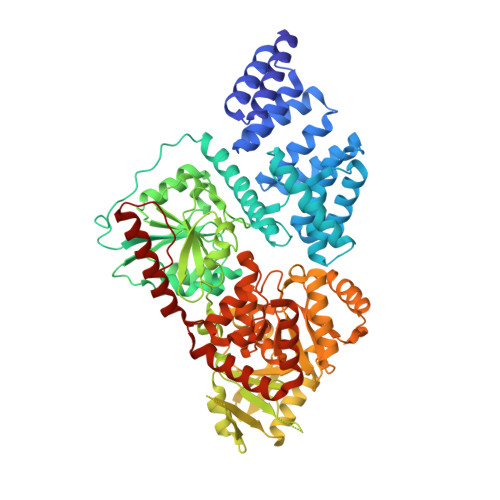
O-Glcnac Transferase Invokes Nucleotide Sugar Pyrophosphate Participation in Catalysis
Schimpl, M., Zheng, X., Borodkin, V.S., Blair, D.E., Ferenbach, A.T., Schuettelkopf, A.W., Navratilova, I., Aristotelous, T., Albarbarawi, O., Robinson, D.A., Macnaughtan, M.A., Van Aalten, D.M.F.(2012) Nat Chem Biol 8: 969
- PubMed: 23103942
- DOI: https://doi.org/10.1038/nchembio.1108
- Primary Citation of Related Structures:
4AY5, 4AY6 - PubMed Abstract:
Protein O-GlcNAcylation is an essential post-translational modification on hundreds of intracellular proteins in metazoa, catalyzed by O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) using unknown mechanisms of transfer and substrate recognition. Through crystallographic snapshots and mechanism-inspired chemical probes, we define how human OGT recognizes the sugar donor and acceptor peptide and uses a new catalytic mechanism of glycosyl transfer, involving the sugar donor α-phosphate as the catalytic base as well as an essential lysine. This mechanism seems to be a unique evolutionary solution to the spatial constraints imposed by a bulky protein acceptor substrate and explains the unexpected specificity of a recently reported metabolic OGT inhibitor.
- College of Life Sciences, University of Dundee, Dundee, UK.
Organizational Affiliation: