Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Liao, H.X., Bonsignori, M., Alam, S.M., McLellan, J.S., Tomaras, G.D., Moody, M.A., Kozink, D.M., Hwang, K.K., Chen, X., Tsao, C.Y., Liu, P., Lu, X., Parks, R.J., Montefiori, D.C., Ferrari, G., Pollara, J., Rao, M., Peachman, K.K., Santra, S., Letvin, N.L., Karasavvas, N., Yang, Z.Y., Dai, K., Pancera, M., Gorman, J., Wiehe, K., Nicely, N.I., Rerks-Ngarm, S., Nitayaphan, S., Kaewkungwal, J., Pitisuttithum, P., Tartaglia, J., Sinangil, F., Kim, J.H., Michael, N.L., Kepler, T.B., Kwong, P.D., Mascola, J.R., Nabel, G.J., Pinter, A., Zolla-Pazner, S., Haynes, B.F.(2013) Immunity 38: 176-186
- PubMed: 23313589
- DOI: https://doi.org/10.1016/j.immuni.2012.11.011
- Primary Citation of Related Structures:
4HPO, 4HPY, 4HQQ - PubMed Abstract:
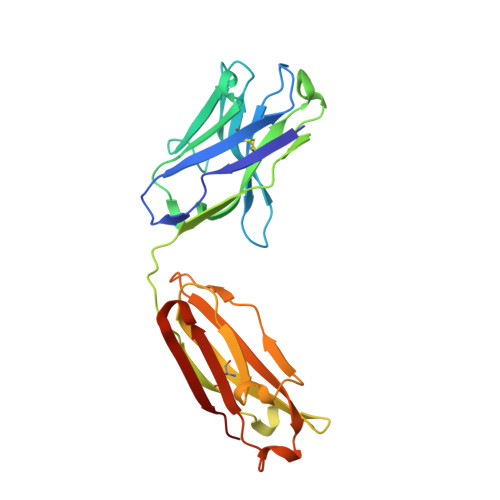
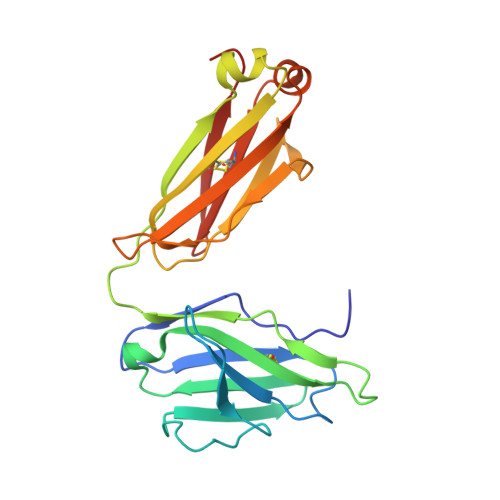
The RV144 HIV-1 trial of the canary pox vector (ALVAC-HIV) plus the gp120 AIDSVAX B/E vaccine demonstrated an estimated efficacy of 31%, which correlated directly with antibodies to HIV-1 envelope variable regions 1 and 2 (V1-V2). Genetic analysis of trial viruses revealed increased vaccine efficacy against viruses matching the vaccine strain at V2 residue 169. Here, we isolated four V2 monoclonal antibodies from RV144 vaccinees that recognize residue 169, neutralize laboratory-adapted HIV-1, and mediate killing of field-isolate HIV-1-infected CD4(+) T cells. Crystal structures of two of the V2 antibodies demonstrated that residue 169 can exist within divergent helical and loop conformations, which contrasted dramatically with the β strand conformation previously observed with a broadly neutralizing antibody PG9. Thus, RV144 vaccine-induced immune pressure appears to target a region that may be both sequence variable and structurally polymorphic. Variation may signal sites of HIV-1 envelope vulnerability, providing vaccine designers with new options.
- Duke Human Vaccine Institute, Duke University Medical Center, Durham, NC 27710, USA. hliao@duke.edu
Organizational Affiliation: