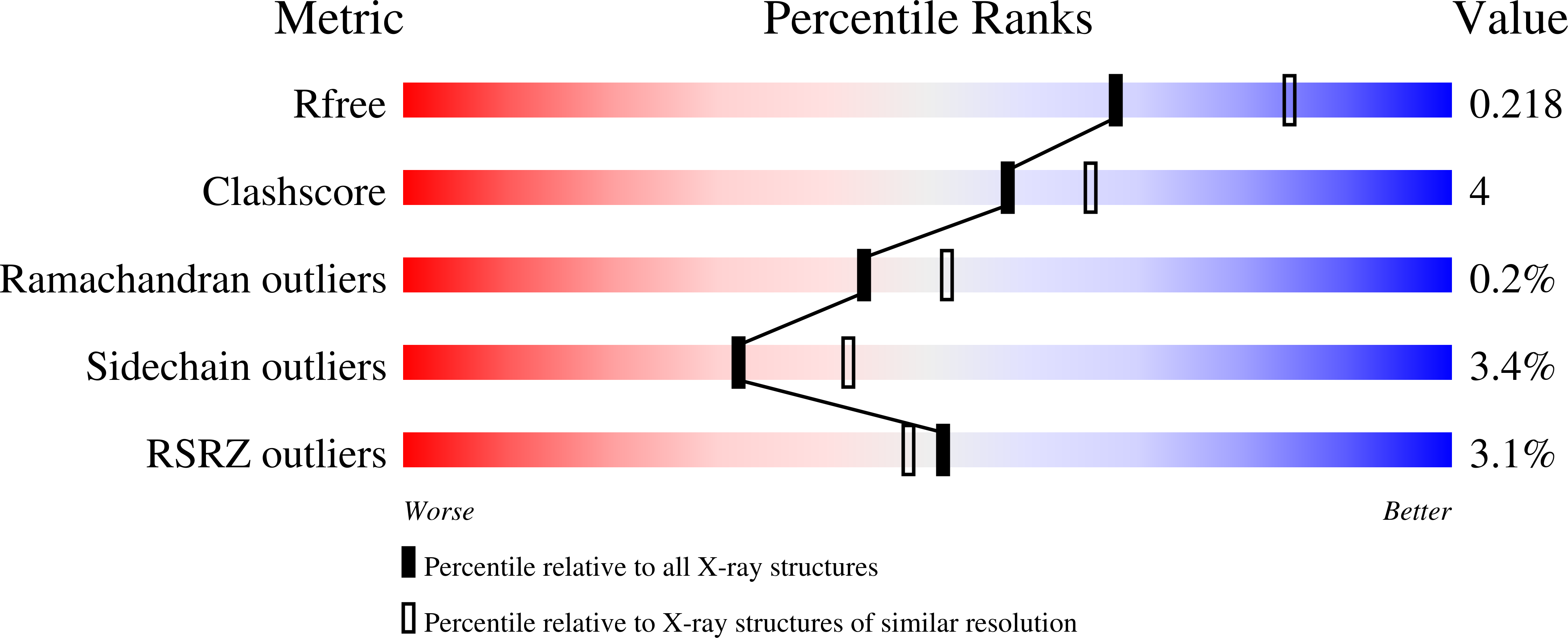
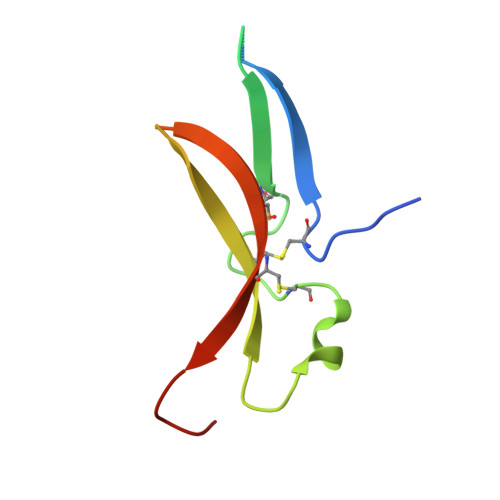
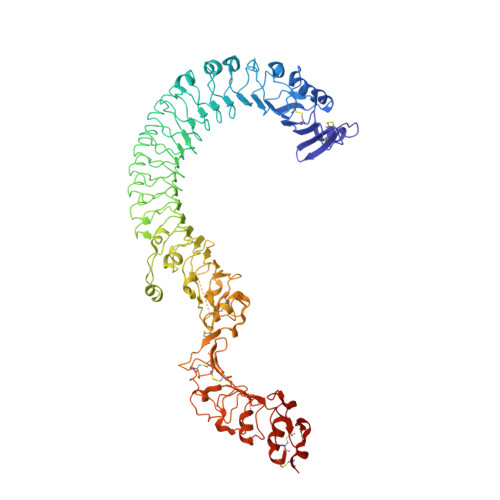Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity.
Parthier, C., Stelter, M., Ursel, C., Fandrich, U., Lilie, H., Breithaupt, C., Stubbs, M.T.(2014) Proc Natl Acad Sci U S A 111: 6281-6286
- PubMed: 24733933
- DOI: https://doi.org/10.1073/pnas.1320678111
- Primary Citation of Related Structures:
4LXR, 4LXS - PubMed Abstract:
Drosophila Toll receptors are involved in embryonic development and the immune response of adult flies. In both processes, the only known Toll receptor ligand is the human nerve growth factor-like cystine knot protein Spätzle. Here we present the crystal structure of a 1:1 (nonsignaling) complex of the full-length Toll receptor ectodomain (ECD) with the Spätzle cystine knot domain dimer. The ECD is divided into two leucine-rich repeat (LRR) domains, each of which is capped by cysteine-rich domains. Spätzle binds to the concave surface of the membrane-distal LRR domain, in contrast to the flanking ligand interactions observed for mammalian Toll-like receptors, with asymmetric contributions from each Spätzle protomer. The structure allows rationalization of existing genetic and biochemical data and provides a framework for targeting the immune systems of insects of economic importance, as well as a variety of invertebrate disease vectors.
- Institut für Biochemie und Biotechnologie and Mitteldeutsches Zentrum für Struktur und Dynamik von Proteinen, Martin-Luther-Universität Halle-Wittenberg, 06120 Halle (Saale), Germany.
Organizational Affiliation: