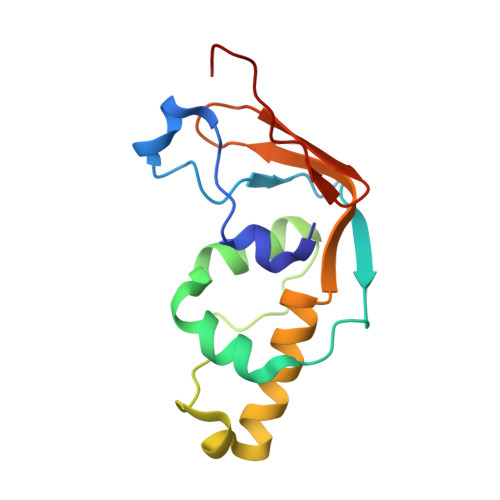
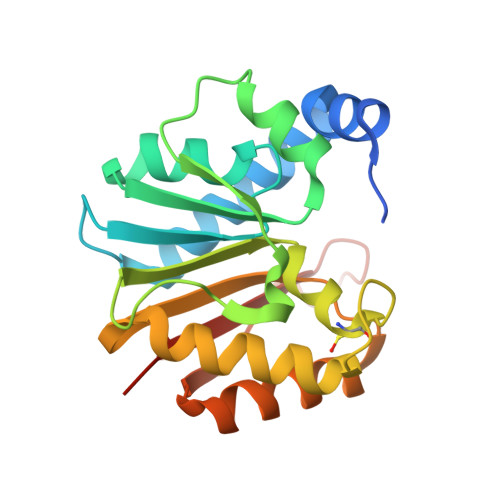
Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes.
Letoquart, J., Huvelle, E., Wacheul, L., Bourgeois, G., Zorbas, C., Graille, M., Heurgue-Hamard, V., Lafontaine, D.L.(2014) Proc Natl Acad Sci U S A 111: E5518-E5526
- PubMed: 25489090
- DOI: https://doi.org/10.1073/pnas.1413089111
- Primary Citation of Related Structures:
4QTT, 4QTU - PubMed Abstract:
The eukaryotic small ribosomal subunit carries only four ribosomal (r) RNA methylated bases, all close to important functional sites. N(7)-methylguanosine (m(7)G) introduced at position 1575 on 18S rRNA by Bud23-Trm112 is at a ridge forming a steric block between P- and E-site tRNAs. Here we report atomic resolution structures of Bud23-Trm112 in the apo and S-adenosyl-L-methionine (SAM)-bound forms. Bud23 and Trm112 interact through formation of a β-zipper involving main-chain atoms, burying an important hydrophobic surface and stabilizing the complex. The structures revealed that the coactivator Trm112 undergoes an induced fit to accommodate its methyltransferase (MTase) partner. We report important structural similarity between the active sites of Bud23 and Coffea canephora xanthosine MTase, leading us to propose and validate experimentally a model for G1575 coordination. We identify Bud23 residues important for Bud23-Trm112 complex formation and recruitment to pre-ribosomes. We report that though Bud23-Trm112 binds precursor ribosomes at an early nucleolar stage, m(7)G methylation occurs at a late step of small subunit biogenesis, implying specifically delayed catalytic activation. Finally, we show that Bud23-Trm112 interacts directly with the box C/D snoRNA U3-associated DEAH RNA helicase Dhr1 supposedly involved in central pseudoknot formation; this suggests that Bud23-Trm112 might also contribute to controlling formation of this irreversible and dramatic structural reorganization essential to overall folding of small subunit rRNA. Our study contributes important new elements to our understanding of key molecular aspects of human ribosomopathy syndromes associated with WBSCR22 (human Bud23) malfunction.
- Laboratoire de Biochimie, CNRS UMR 7654, Ecole Polytechnique, F-91128 Palaiseau Cedex, France;
Organizational Affiliation: