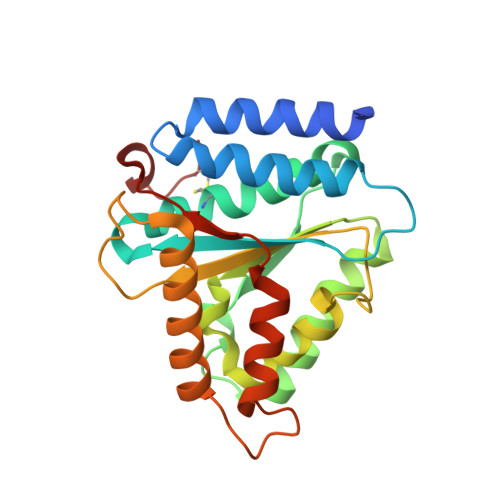
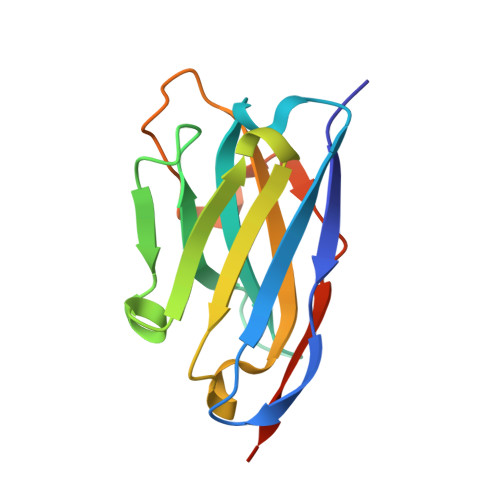
How lamina-associated polypeptide 1 (LAP1) activates Torsin.
Sosa, B.A., Demircioglu, F.E., Chen, J.Z., Ingram, J., Ploegh, H.L., Schwartz, T.U.(2014) Elife 3: e03239-e03239
- PubMed: 25149450
- DOI: https://doi.org/10.7554/eLife.03239
- Primary Citation of Related Structures:
4TVS - PubMed Abstract:
Lamina-associated polypeptide 1 (LAP1) resides at the nuclear envelope and interacts with Torsins, poorly understood endoplasmic reticulum (ER)-localized AAA+ ATPases, through a conserved, perinuclear domain. We determined the crystal structure of the perinuclear domain of human LAP1. LAP1 possesses an atypical AAA+ fold. While LAP1 lacks canonical nucleotide binding motifs, its strictly conserved arginine 563 is positioned exactly where the arginine finger of canonical AAA+ ATPases is found. Based on modeling and electron microscopic analysis, we propose that LAP1 targets Torsin to the nuclear envelope by forming an alternating, heterohexameric (LAP1-Torsin)3 ring, in which LAP1 acts as the Torsin activator. The experimental data show that mutation of arginine 563 in LAP1 reduces its ability to stimulate TorsinA ATPase hydrolysis. This knowledge may help scientists understand the etiology of DYT1 primary dystonia, a movement disorder caused by a single glutamate deletion in TorsinA.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, United States.
Organizational Affiliation: