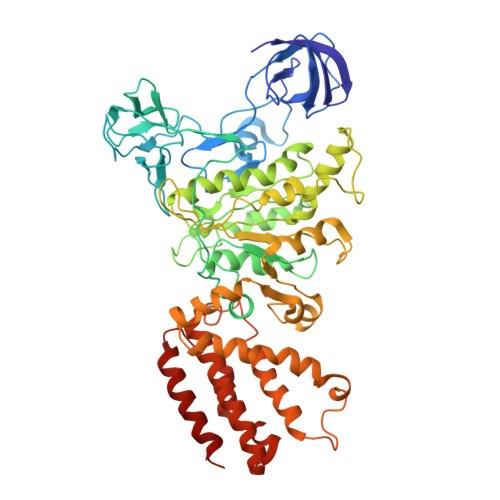
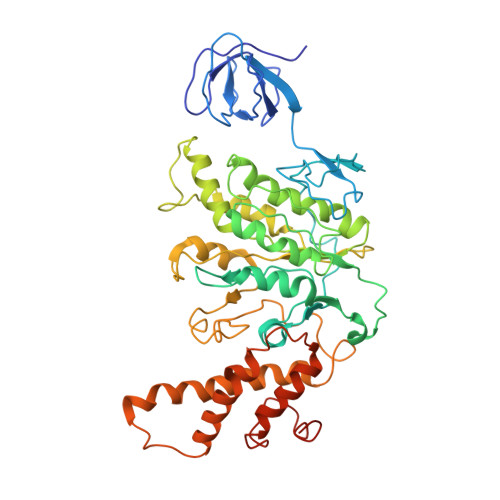
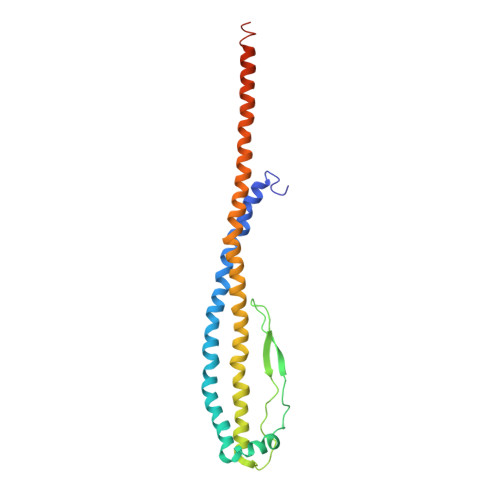
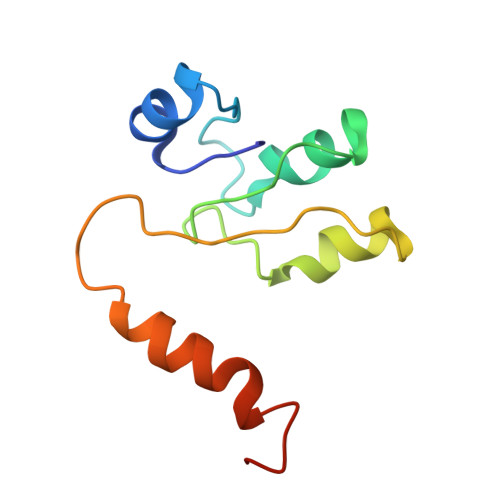
Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nakanishi, A., Kishikawa, J.I., Tamakoshi, M., Mitsuoka, K., Yokoyama, K.(2018) Nat Commun 9: 89-89
- PubMed: 29311594
- DOI: https://doi.org/10.1038/s41467-017-02553-6
- Primary Citation of Related Structures:
5Y5X, 5Y5Y, 5Y5Z, 5Y60 - PubMed Abstract:
Proton translocating rotary ATPases couple ATP hydrolysis/synthesis, which occurs in the soluble domain, with proton flow through the membrane domain via a rotation of the common central rotor complex against the surrounding peripheral stator apparatus. Here, we present a large data set of single particle cryo-electron micrograph images of the V/A type H + -rotary ATPase from the bacterium Thermus thermophilus, enabling the identification of three rotational states based on the orientation of the rotor subunit. Using masked refinement and classification with signal subtractions, we obtain homogeneous reconstructions for the whole complexes and soluble V 1 domains. These reconstructions are of higher resolution than any EM map of intact rotary ATPase reported previously, providing a detailed molecular basis for how the rotary ATPase maintains structural integrity of the peripheral stator apparatus, and confirming the existence of a clear proton translocation path from both sides of the membrane.
- Department of Molecular Biosciences, Kyoto Sangyo University, Motoyama Kamigamo, Kita-ku, Kyoto, 603-8555, Japan.
Organizational Affiliation: