Biochemical and Structural Insights into the Preference of Nairoviral DeISGylases for Interferon-Stimulated Gene Product 15 Originating from Certain Species.
Deaton, M.K., Dzimianski, J.V., Daczkowski, C.M., Whitney, G.K., Mank, N.J., Parham, M.M., Bergeron, E., Pegan, S.D.(2016) J Virol 90: 8314-8327
- PubMed: 27412597
- DOI: https://doi.org/10.1128/JVI.00975-16
- Primary Citation of Related Structures:
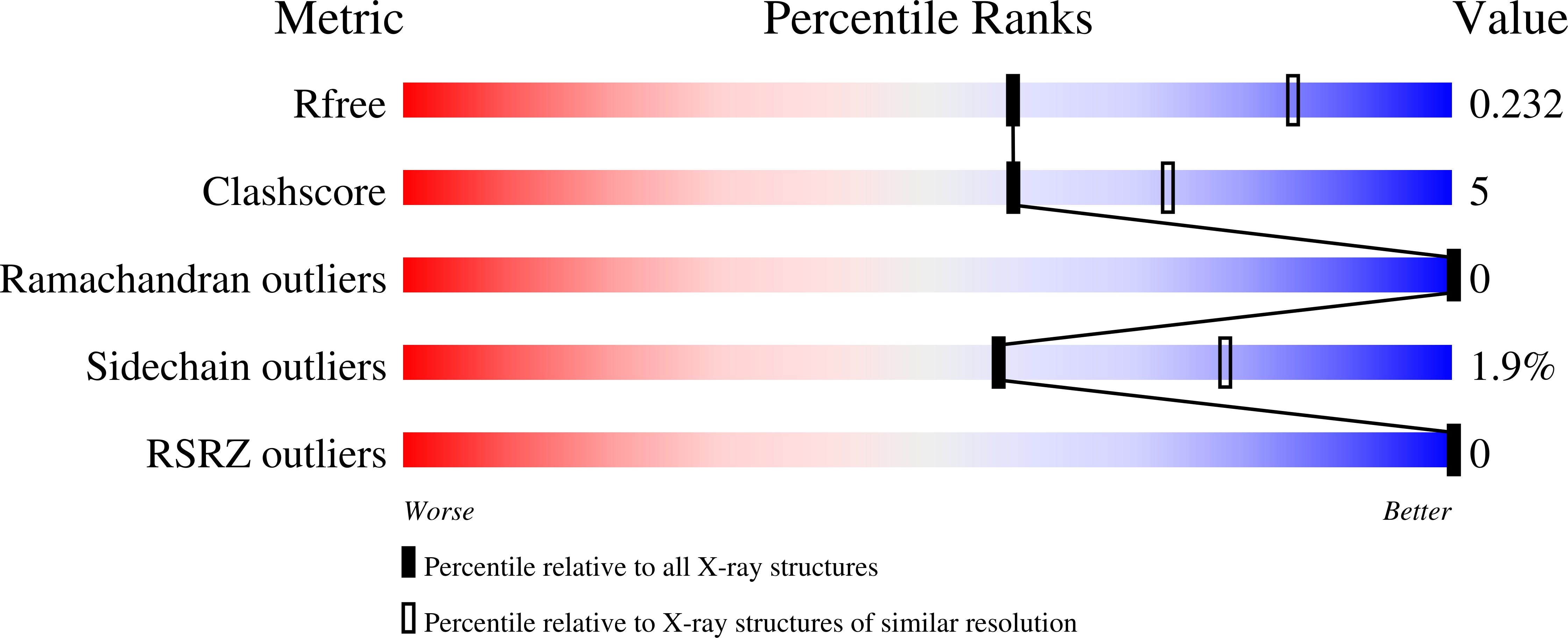
5JZE - PubMed Abstract:
The regulation of the interferon type I (IFN-I) response has been shown to rely on posttranslational modification by ubiquitin (Ub) and Ub-like interferon-stimulated gene product 15 (ISG15) to stabilize, or activate, a variety of IFN-I signaling and downstream effector proteins. Unlike Ub, which is almost perfectly conserved among eukaryotes, ISG15 is highly divergent, even among mammals. Since zoonotic viruses rely on viral proteins to recognize, or cleave, ISG15 conjugates in order to evade, or suppress, innate immunity, the impact of ISG15 biodiversity on deISGylating proteases of the ovarian tumor family (vOTU) from nairoviruses was evaluated. The enzymatic activities of vOTUs originating from the Crimean-Congo hemorrhagic fever virus, Erve virus, and Nairobi sheep disease virus were tested against ISG15s from humans, mice, shrews, sheep, bats, and camels, which are mammalian species known to be infected by nairoviruses. This along with investigation of binding by isothermal titration calorimetry illustrated significant differences in the abilities of nairovirus deISGylases to accommodate certain species of ISG15. To investigate the molecular underpinnings of species preferences of these vOTUs, a structure was determined to 2.5 Å for a complex of Erve virus vOTU protease and a mouse ISG15 domain. This structure revealed the molecular basis of Erve virus vOTU's preference for ISG15 over Ub and the first structural insight into a nonhuman ISG15. This structure also revealed key interactions, or lack thereof, surrounding three amino acids that may drive a viral deISgylase to prefer an ISG15 from one species over that of another. Viral ovarian tumor domain proteases (vOTUs) are one of the two principal classes of viral proteases observed to reverse posttranslational modification of host proteins by ubiquitin and interferon-stimulated gene product 15 (ISG15), subsequently facilitating downregulation of IFN-I signaling pathways. Unlike the case with ubiquitin, the amino acid sequences of ISG15s from various species are notably divergent. We illustrate that vOTUs have clear preferences for ISG15s from certain species. In addition, these observations are related to the molecular insights acquired via the first X-ray structure of the vOTU from the Erve nairovirus in complex with the first structurally resolved nonhuman ISG15. This information implicates certain amino acids that drive the preference of vOTUs for ISG15s from certain species.
Organizational Affiliation:
Department of Pharmaceutical and Biomedical Sciences, University of Georgia, Athens, Georgia, USA.