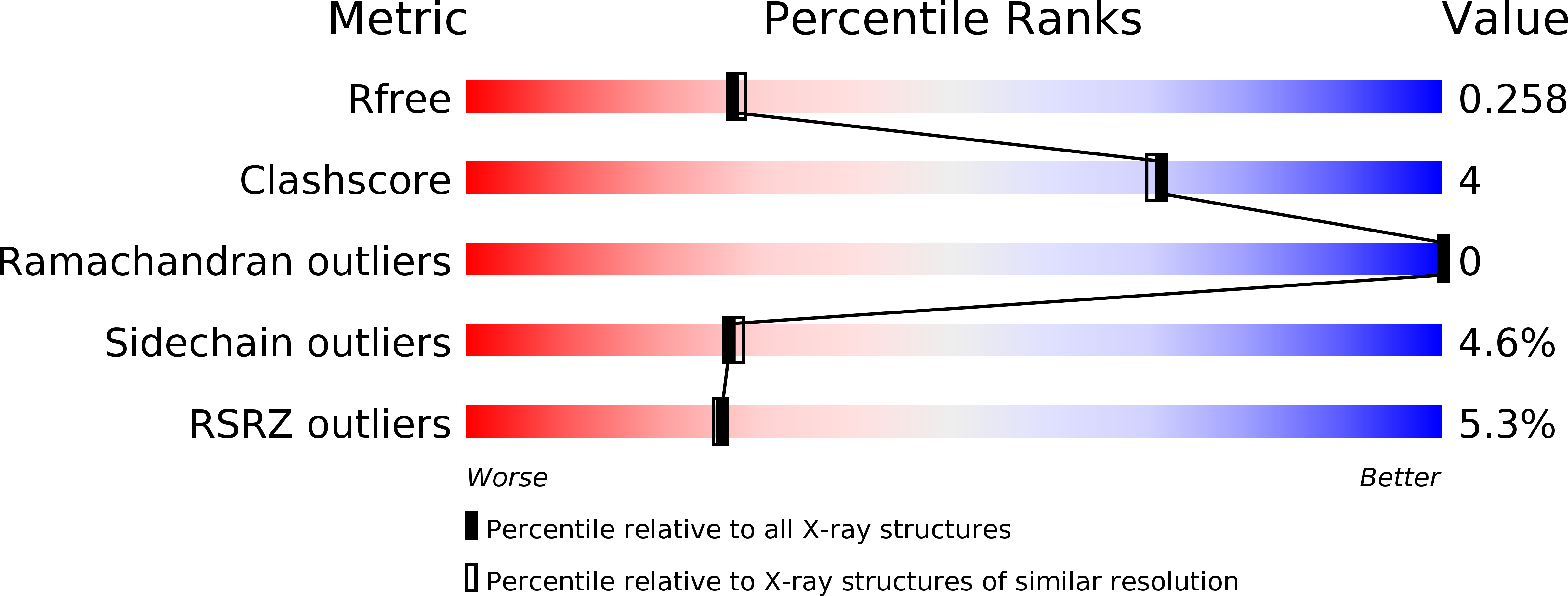
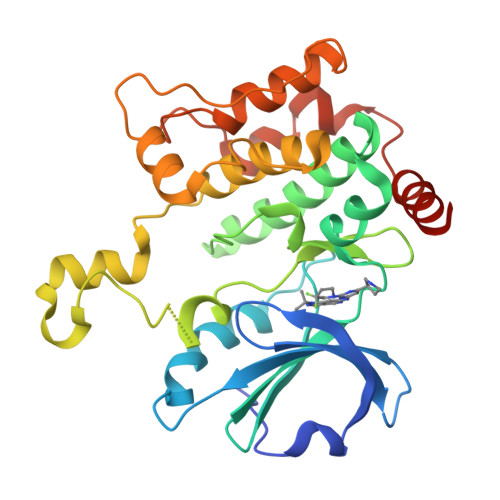
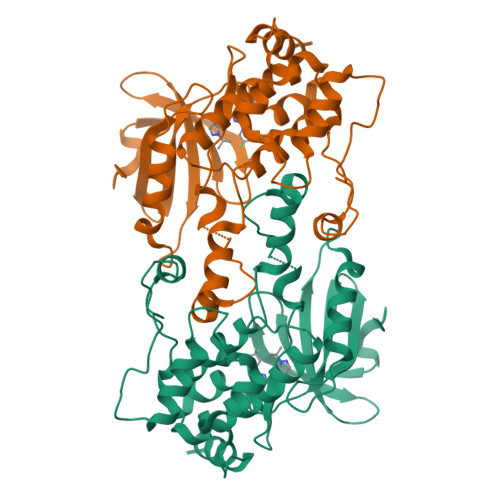
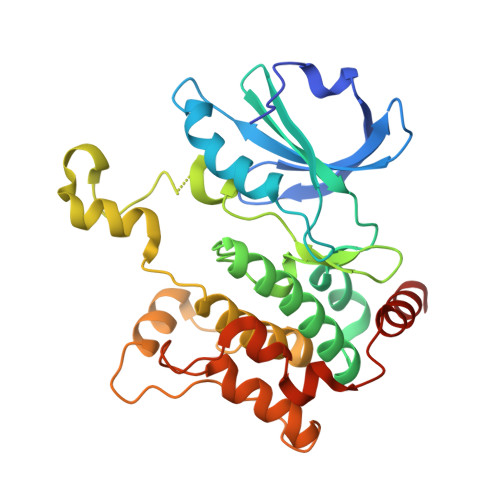
Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Wu, P., Sneeringer, C.J., Pitts, K.E., Day, E.S., Chan, B.K., Wei, B., Lehoux, I., Mortara, K., Li, H., Wu, J., Franke, Y., Moffat, J.G., Grogan, J.L., Heffron, T.P., Wang, W.(2019) Structure 27: 125-133.e4
- PubMed: 30503777
- DOI: https://doi.org/10.1016/j.str.2018.10.025
- Primary Citation of Related Structures:
6CQD, 6CQE, 6CQF - PubMed Abstract:
Enhancement of antigen-specific T cell immunity has shown significant therapeutic benefit in infectious diseases and cancer. Hematopoietic progenitor kinase-1 (HPK1) is a negative-feedback regulator of T cell receptor signaling, which dampens T cell proliferation and effector function. A recent report showed that a catalytic dead mutant of HPK1 phenocopies augmented T cell responses observed in HPK1-knockout mice, indicating that kinase activity is critical for function. We evaluated active and inactive mutants and determined crystal structures of HPK1 kinase domain (HPK1-KD) in apo and ligand bound forms. In all structures HPK1-KD displays a rare domain-swapped dimer, in which the activation segment comprises a well-conserved dimer interface. Biophysical measurements show formation of dimer in solution. The activation segment adopts an α-helical structure which exhibits distinct orientations in active and inactive states. This face-to-face configuration suggests that the domain-swapped dimer may possess alternative selectivity for certain substrates of HPK1 under relevant cellular context.
Organizational Affiliation:
Department of Structural Biology, Genentech, South San Francisco, CA 94080, USA.