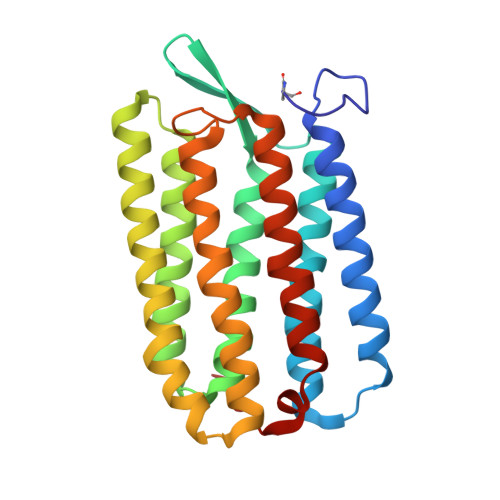
Two states of a light-sensitive membrane protein captured at room temperature using thin-film sample mounts.
Axford, D., Judge, P.J., Bada Juarez, J.F., Kwan, T.O.C., Birch, J., Vinals, J., Watts, A., Moraes, I.(2022) Acta Crystallogr D Struct Biol 78: 52-58
- PubMed: 34981761
- DOI: https://doi.org/10.1107/S2059798321011220
- Primary Citation of Related Structures:
6S63 - PubMed Abstract:
Room-temperature diffraction methods are highly desirable for dynamic studies of biological macromolecules, since they allow high-resolution structural data to be collected as proteins undergo conformational changes. For crystals grown in lipidic cubic phase (LCP), an extruder is commonly used to pass a stream of microcrystals through the X-ray beam; however, the sample quantities required for this method may be difficult to produce for many membrane proteins. A more sample-efficient environment was created using two layers of low X-ray transmittance polymer films to mount crystals of the archaerhodopsin-3 (AR3) photoreceptor and room-temperature diffraction data were acquired. By using transparent and opaque polymer films, two structures, one corresponding to the desensitized, dark-adapted (DA) state and the other to the ground or light-adapted (LA) state, were solved to better than 1.9 Å resolution. All of the key structural features of AR3 were resolved, including the retinal chromophore, which is present as the 13-cis isomer in the DA state and as the all-trans isomer in the LA state. The film-sandwich sample environment enables diffraction data to be recorded at room temperature in both illuminated and dark conditions, which more closely approximate those in vivo. This simple approach is applicable to a wide range of membrane proteins crystallized in LCP and light-sensitive samples in general at synchrotron and laboratory X-ray sources.
- Diamond Light Source, Harwell Science and Innovation Campus, Didcot OX11 0DE, United Kingdom.
Organizational Affiliation: