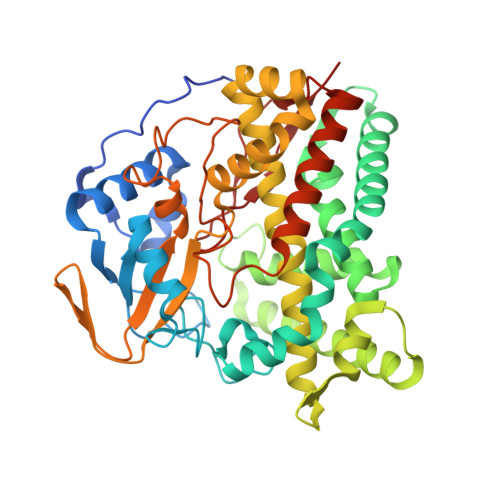
Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
Varaksa, T., Bukhdruker, S., Grabovec, I., Marin, E., Kavaleuski, A., Gusach, A., Kovalev, K., Maslov, I., Luginina, A., Zabelskii, D., Astashkin, R., Shevtsov, M., Smolskaya, S., Kavaleuskaya, A., Shabunya, P., Baranovsky, A., Dolgopalets, V., Charnou, Y., Savachka, A., Litvinovskaya, R., Hurski, A., Shevchenko, E., Rogachev, A., Mishin, A., Gordeliy, V., Gabrielian, A., Hurt, D.E., Nikonenko, B., Majorov, K., Apt, A., Rosenthal, A., Gilep, A., Borshchevskiy, V., Strushkevich, N.(2021) J Mol Biology 433: 166763-166763
- PubMed: 33359098
- DOI: https://doi.org/10.1016/j.jmb.2020.166763
- Primary Citation of Related Structures:
6T0F, 6T0G, 6T0H, 6T0K, 6T0L - PubMed Abstract:
Mycobacterium tuberculosis (Mtb) infection is among top ten causes of death worldwide, and the number of drug-resistant strains is increasing. The direct interception of human immune signaling molecules by Mtb remains elusive, limiting drug discovery. Oxysterols and secosteroids regulate both innate and adaptive immune responses. Here we report a functional, structural, and bioinformatics study of Mtb enzymes initiating cholesterol catabolism and demonstrated their interrelation with human immunity. We show that these enzymes metabolize human immune oxysterol messengers. Rv2266 - the most potent among them - can also metabolize vitamin D3 (VD3) derivatives. High-resolution structures show common patterns of sterols binding and reveal a site for oxidative attack during catalysis. Finally, we designed a compound that binds and inhibits three studied proteins. The compound shows activity against Mtb H37Rv residing in macrophages. Our findings contribute to molecular understanding of suppression of immunity and suggest that Mtb has its own transformation system resembling the human phase I drug-metabolizing system.
- Institute of Bioorganic Chemistry, National Academy of Sciences of Belarus, Minsk, Belarus.
Organizational Affiliation: