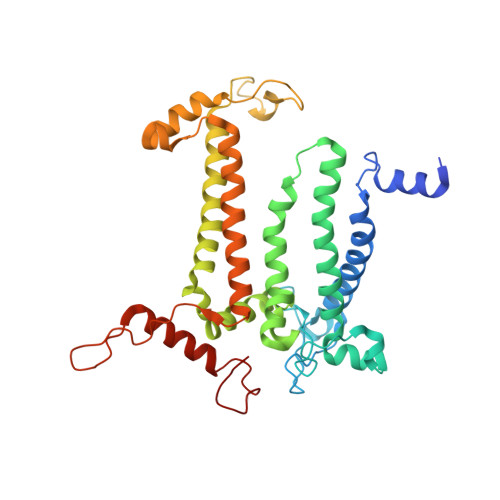
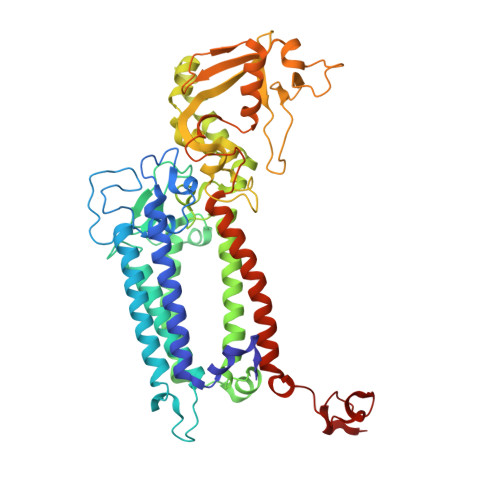
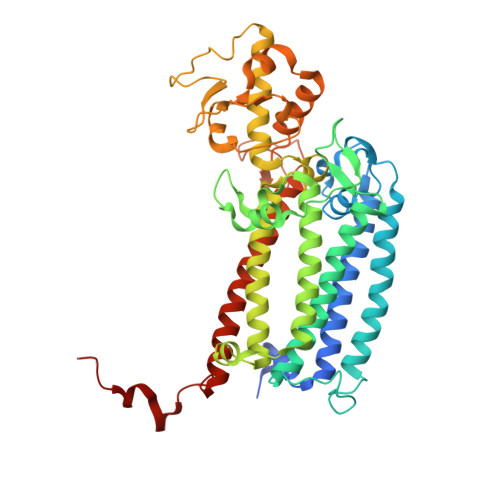
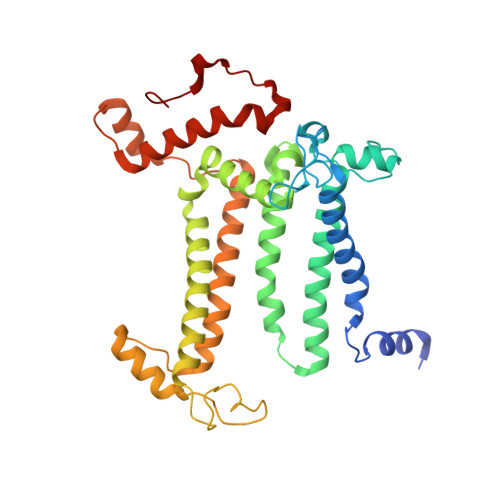
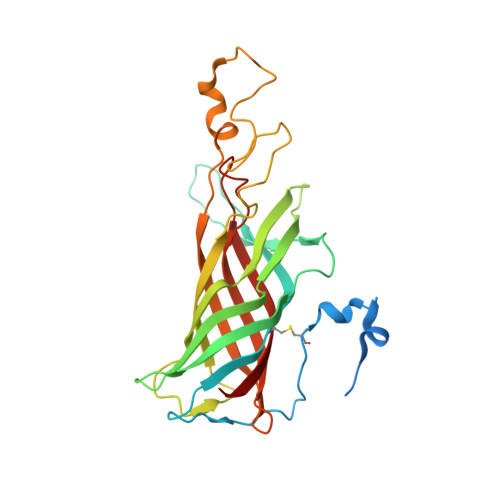
Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Ibrahim, M., Fransson, T., Chatterjee, R., Cheah, M.H., Hussein, R., Lassalle, L., Sutherlin, K.D., Young, I.D., Fuller, F.D., Gul, S., Kim, I.S., Simon, P.S., de Lichtenberg, C., Chernev, P., Bogacz, I., Pham, C.C., Orville, A.M., Saichek, N., Northen, T., Batyuk, A., Carbajo, S., Alonso-Mori, R., Tono, K., Owada, S., Bhowmick, A., Bolotovsky, R., Mendez, D., Moriarty, N.W., Holton, J.M., Dobbek, H., Brewster, A.S., Adams, P.D., Sauter, N.K., Bergmann, U., Zouni, A., Messinger, J., Kern, J., Yachandra, V.K., Yano, J.(2020) Proc Natl Acad Sci U S A 117: 12624-12635
- PubMed: 32434915
- DOI: https://doi.org/10.1073/pnas.2000529117
- Primary Citation of Related Structures:
6W1O, 6W1P, 6W1Q, 6W1R, 6W1T, 6W1U, 6W1V - PubMed Abstract:
In oxygenic photosynthesis, light-driven oxidation of water to molecular oxygen is carried out by the oxygen-evolving complex (OEC) in photosystem II (PS II). Recently, we reported the room-temperature structures of PS II in the four (semi)stable S-states, S 1 , S 2 , S 3 , and S 0 , showing that a water molecule is inserted during the S 2 → S 3 transition, as a new bridging O(H)-ligand between Mn1 and Ca. To understand the sequence of events leading to the formation of this last stable intermediate state before O 2 formation, we recorded diffraction and Mn X-ray emission spectroscopy (XES) data at several time points during the S 2 → S 3 transition. At the electron acceptor site, changes due to the two-electron redox chemistry at the quinones, Q A and Q B , are observed. At the donor site, tyrosine Y Z and His190 H-bonded to it move by 50 µs after the second flash, and Glu189 moves away from Ca. This is followed by Mn1 and Mn4 moving apart, and the insertion of O X (H) at the open coordination site of Mn1. This water, possibly a ligand of Ca, could be supplied via a "water wheel"-like arrangement of five waters next to the OEC that is connected by a large channel to the bulk solvent. XES spectra show that Mn oxidation (τ of ∼350 µs) during the S 2 → S 3 transition mirrors the appearance of O X electron density. This indicates that the oxidation state change and the insertion of water as a bridging atom between Mn1 and Ca are highly correlated.
- Institut für Biologie, Humboldt-Universität zu Berlin, D-10115 Berlin, Germany.
Organizational Affiliation: