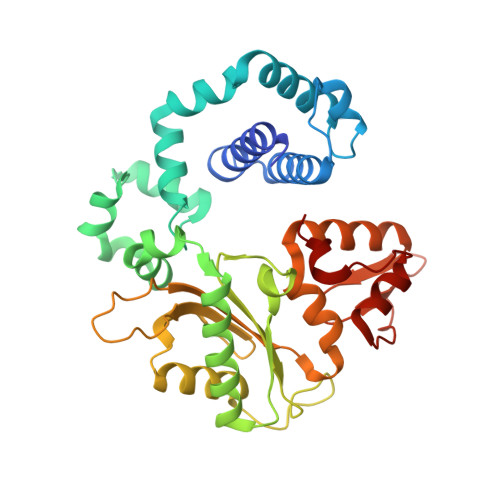
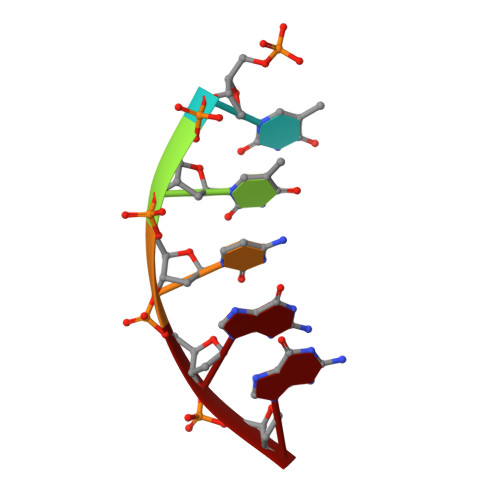
Revealing an Internal Stabilization Deficiency in the DNA Polymerase beta K289M Cancer Variant through the Combined Use of Chemical Biology and X-ray Crystallography.
Batra, V.K., Alnajjar, K.S., Sweasy, J.B., McKenna, C.E., Goodman, M.F., Wilson, S.H.(2020) Biochemistry 59: 955-963
- PubMed: 31999437
- DOI: https://doi.org/10.1021/acs.biochem.9b01072
- Primary Citation of Related Structures:
6NKR, 6NKS, 6NKT, 6NKU, 6NKV, 6NKW, 6NKX, 6NKY, 6NKZ, 6NL0 - PubMed Abstract:
The human DNA polymerase (pol) β cancer variant K289M has altered polymerase activity in vitro , and the structure of wild-type pol β reveals that the K289 side chain contributes to a network of stabilizing interactions in a C-terminal region of the enzyme distal to the active site. Here, we probed the capacity of the K289M variant to tolerate strain introduced within the C-terminal region and active site. Strain was imposed by making use of a dGTP analogue containing a CF 2 group substitution for the β-γ bridging oxygen atom. The ternary complex structure of the K289M variant displays an alteration in the C-terminal region, whereas the structure of wild-type pol β is not altered in the presence of the dGTP CF 2 analogue. The alteration in the K289M variant impacts the active site, because the enzyme in the ternary complex fails to adopt the normal open to closed conformational change and assembly of the catalytically competent active site. These results reveal the importance of the K289-mediated stabilizing network in the C-terminal region of pol β and suggest an explanation for why the K289M cancer variant is deficient in polymerase activity even though the position 289 side chain is distal to the active site.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, 111 T. W. Alexander Drive, Research Triangle Park, North Carolina 27709, United States.