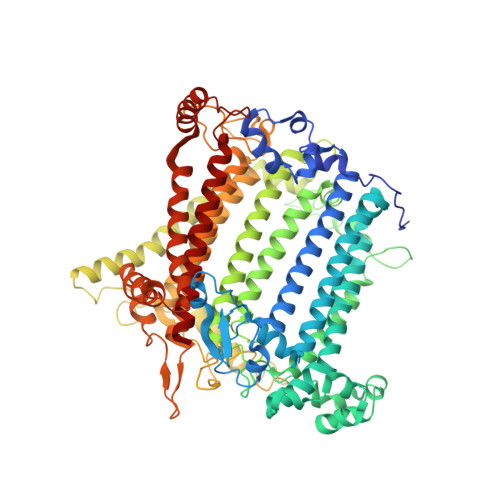
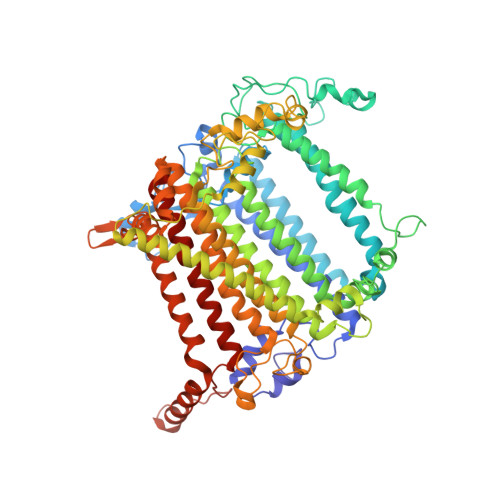
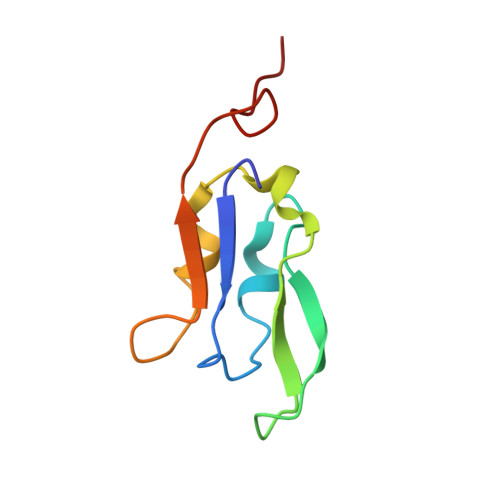
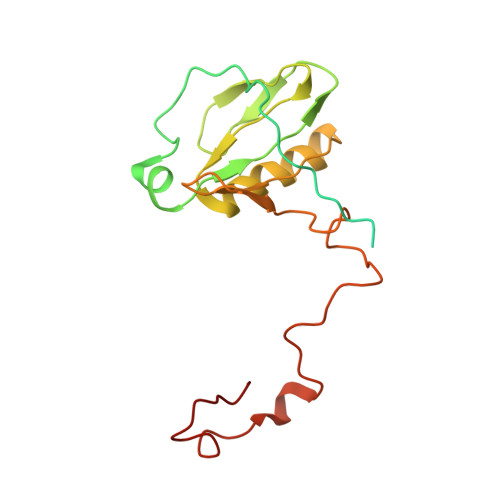
Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Shen, L., Tang, K., Wang, W., Wang, C., Wu, H., Mao, Z., An, S., Chang, S., Kuang, T., Shen, J.R., Han, G., Zhang, X.(2022) Nature 601: 649-654
- PubMed: 34879391
- DOI: https://doi.org/10.1038/s41586-021-04277-6
- Primary Citation of Related Structures:
7EU3, 7EW6, 7EWK, 7F9O - PubMed Abstract:
The chloroplast NADH dehydrogenase-like (NDH) complex is composed of at least 29 subunits and has an important role in mediating photosystem I (PSI) cyclic electron transport (CET) 1-3 . The NDH complex associates with PSI to form the PSI-NDH supercomplex and fulfil its function. Here, we report cryo-electron microscopy structures of a PSI-NDH supercomplex from barley (Hordeum vulgare). The structures reveal that PSI-NDH is composed of two copies of the PSI-light-harvesting complex I (LHCI) subcomplex and one NDH complex. Two monomeric LHCI proteins, Lhca5 and Lhca6, mediate the binding of two PSI complexes to NDH. Ten plant chloroplast-specific NDH subunits are presented and their exact positions as well as their interactions with other subunits in NDH are elucidated. In all, this study provides a structural basis for further investigations on the functions and regulation of PSI-NDH-dependent CET.
Organizational Affiliation:
Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China.