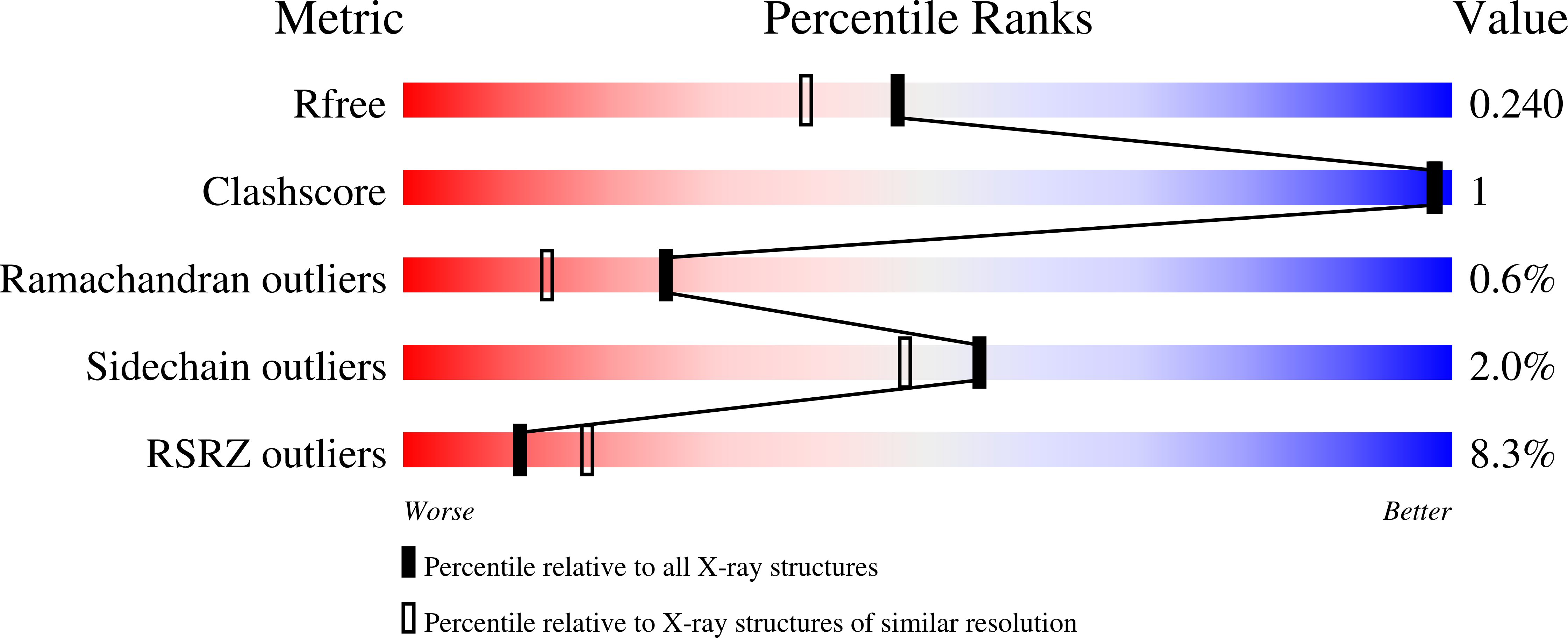
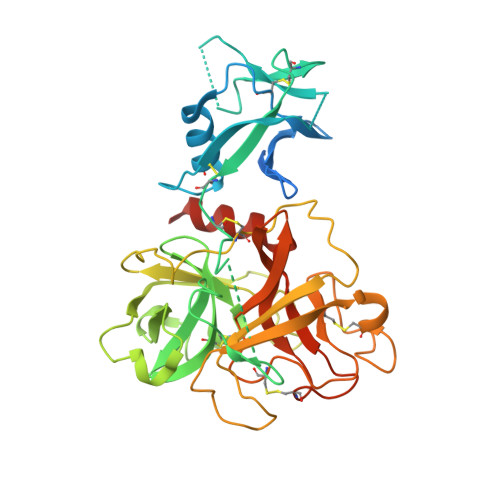
Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Fraser, B.J., Beldar, S., Seitova, A., Hutchinson, A., Mannar, D., Li, Y., Kwon, D., Tan, R., Wilson, R.P., Leopold, K., Subramaniam, S., Halabelian, L., Arrowsmith, C.H., Benard, F.(2022) Nat Chem Biol 18: 963-971
- PubMed: 35676539
- DOI: https://doi.org/10.1038/s41589-022-01059-7
- Primary Citation of Related Structures:
7MEQ - PubMed Abstract:
Transmembrane protease, serine 2 (TMPRSS2) has been identified as key host cell factor for viral entry and pathogenesis of SARS-CoV-2. Specifically, TMPRSS2 proteolytically processes the SARS-CoV-2 Spike (S) protein, enabling virus-host membrane fusion and infection of the airways. We present here a recombinant production strategy for enzymatically active TMPRSS2 and characterization of its matured proteolytic activity, as well as its 1.95 Å X-ray cocrystal structure with the synthetic protease inhibitor nafamostat. Our study provides a structural basis for the potent but nonspecific inhibition by nafamostat and identifies distinguishing features of the TMPRSS2 substrate binding pocket that explain specificity. TMPRSS2 cleaved SARS-CoV-2 S protein at multiple sites, including the canonical S1/S2 cleavage site. We ranked the potency of clinical protease inhibitors with half-maximal inhibitory concentrations ranging from 1.4 nM to 120 µM and determined inhibitor mechanisms of action, providing the groundwork for drug development efforts to selectively inhibit TMPRSS2.
Organizational Affiliation:
Department of Molecular Oncology, British Columbia Cancer Research Institute, Vancouver, British Columbia, Canada.