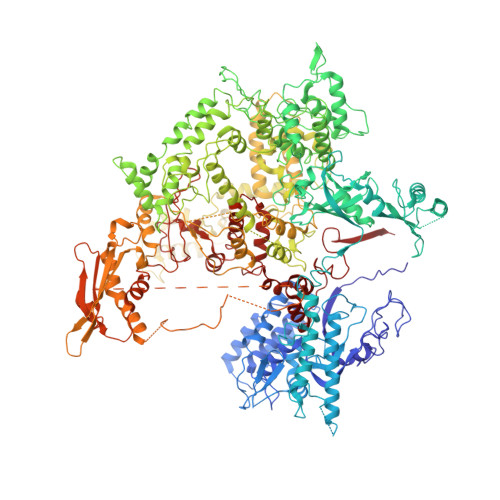
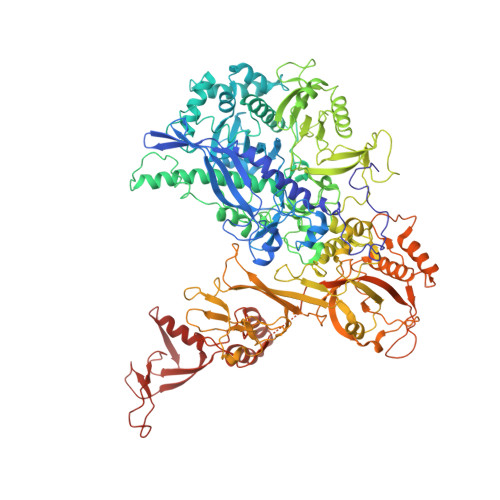
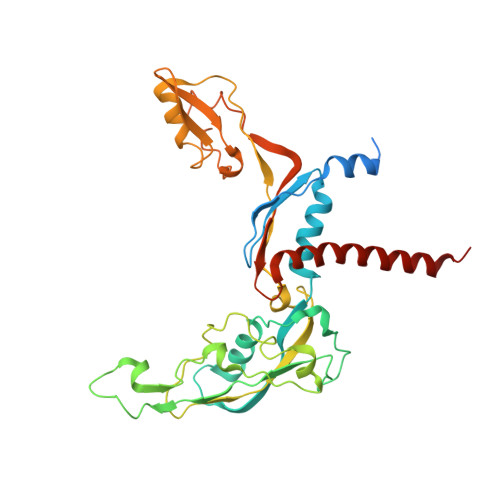
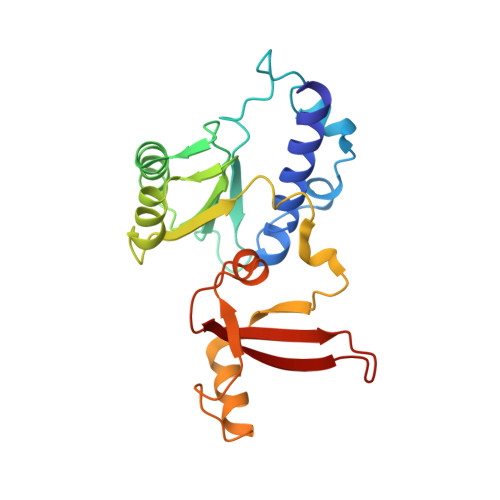
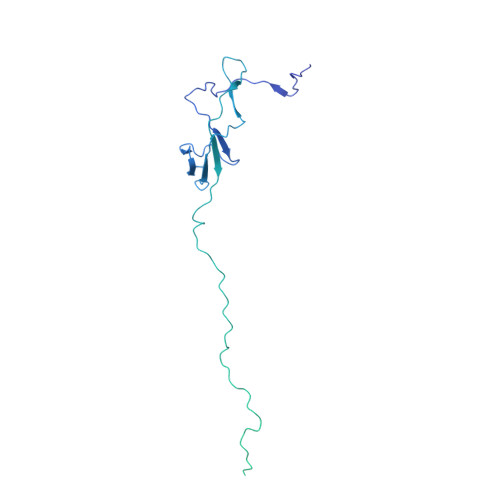
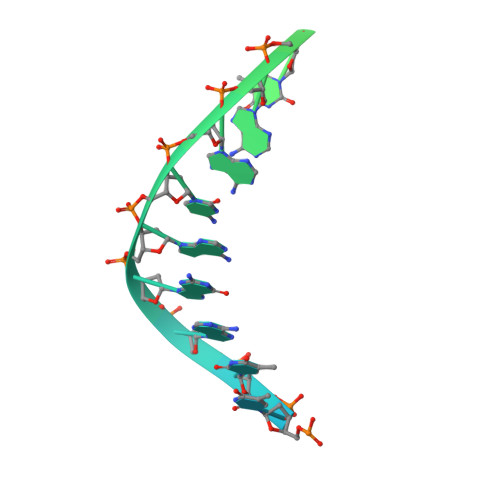
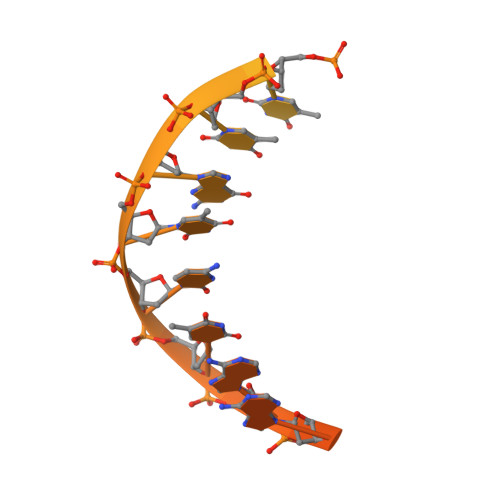
Cryo-EM structures of human RNA polymerase I.
Misiaszek, A.D., Girbig, M., Grotsch, H., Baudin, F., Murciano, B., Lafita, A., Muller, C.W.(2021) Nat Struct Mol Biol 28: 997-1008
- PubMed: 34887565
- DOI: https://doi.org/10.1038/s41594-021-00693-4
- Primary Citation of Related Structures:
7OB9, 7OBA, 7OBB - PubMed Abstract:
RNA polymerase I (Pol I) specifically synthesizes ribosomal RNA. Pol I upregulation is linked to cancer, while mutations in the Pol I machinery lead to developmental disorders. Here we report the cryo-EM structure of elongating human Pol I at 2.7 Å resolution. In the exit tunnel, we observe a double-stranded RNA helix that may support Pol I processivity. Our structure confirms that human Pol I consists of 13 subunits with only one subunit forming the Pol I stalk. Additionally, the structure of human Pol I in complex with the initiation factor RRN3 at 3.1 Å resolution reveals stalk flipping upon RRN3 binding. We also observe an inactivated state of human Pol I bound to an open DNA scaffold at 3.3 Å resolution. Lastly, the high-resolution structure of human Pol I allows mapping of disease-related mutations that can aid understanding of disease etiology.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany.
Organizational Affiliation: