Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
Ryan, B.J., Yang, H., Bacurio, J.H.T., Smith, M.R., Basu, A.K., Greenberg, M.M., Freudenthal, B.D.(2022) J Am Chem Soc 144: 8054-8065
- PubMed: 35499923
- DOI: https://doi.org/10.1021/jacs.2c00193
- Primary Citation of Related Structures:
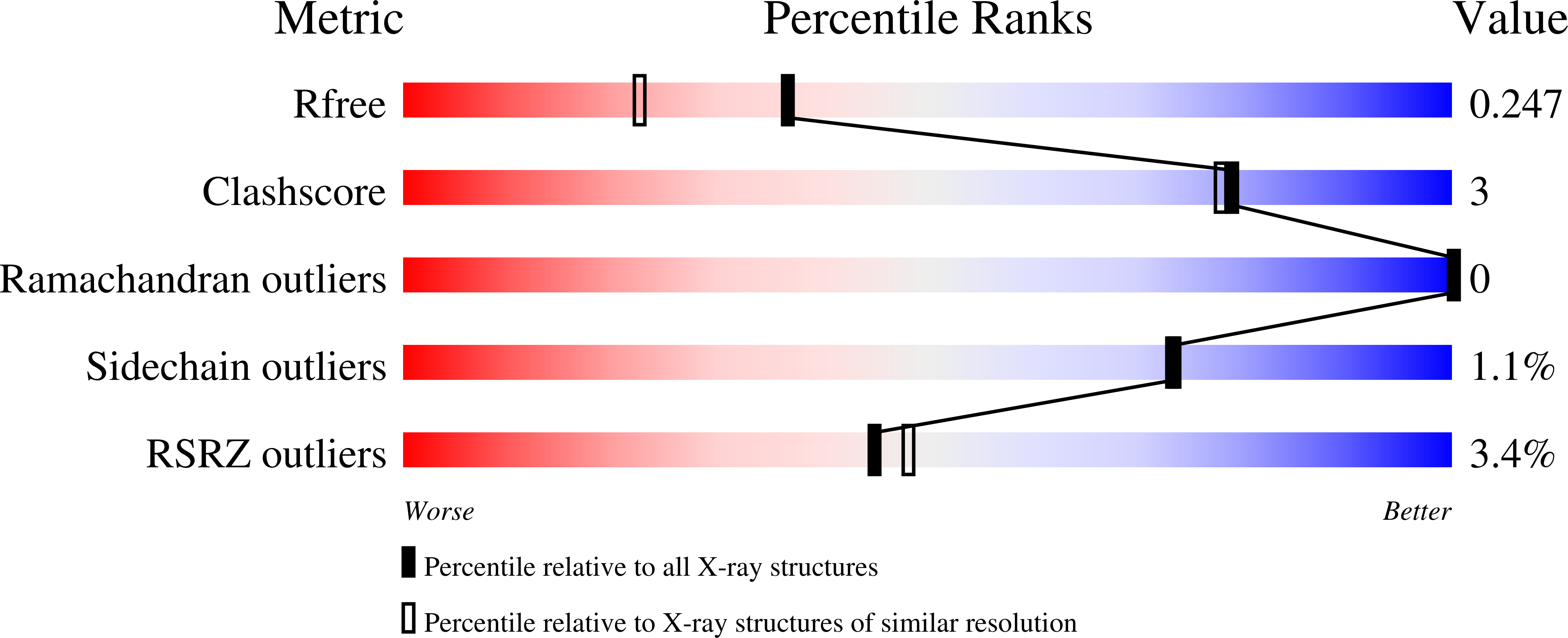
7S9J, 7S9K, 7S9L, 7S9M, 7S9N, 7S9O, 7S9P, 7S9Q - PubMed Abstract:
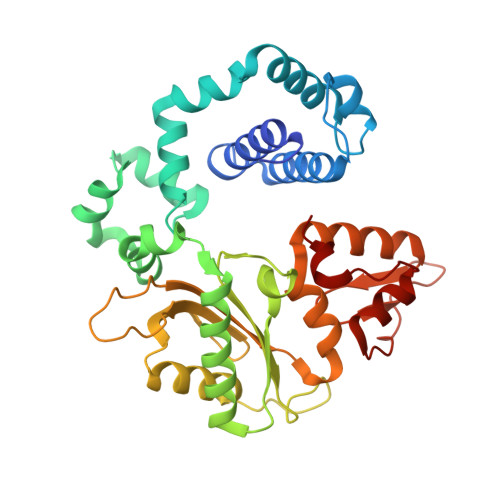
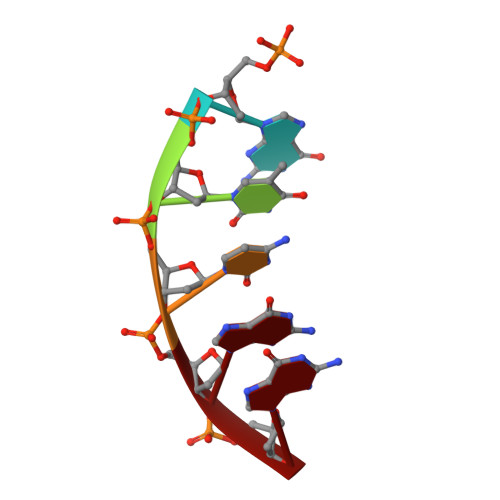
N 6-(2-Deoxy-α,β-d- erythro -pentofuranosyl)-2,6-diamino-4-hydroxy-5-formamido pyrimidine (Fapy•dG) is a prevalent form of genomic DNA damage. Fapy•dG is formed in greater amounts under anoxic conditions than the well-studied, chemically related 7,8-dihydro-8-oxo-2'-deoxyguanosine (8-oxodGuo). Fapy•dG is more mutagenic in mammalian cells than 8-oxodGuo. A distinctive property of Fapy•dG is facile epimerization, but prior works with Fapy•dG analogues have precluded determining its effect on chemistry. We present crystallographic characterization of natural Fapy•dG in duplex DNA and as the template base for DNA polymerase β (Pol β). Fapy•dG adopts the β-anomer when base paired with cytosine but exists as a mixture of α- and β-anomers when promutagenically base paired with adenine. Rotation about the bond between the glycosidic nitrogen atom and the pyrimidine ring is also affected by the opposing nucleotide. Sodium cyanoborohydride soaking experiments trap the ring-opened Fapy•dG, demonstrating that ring opening and epimerization occur in the crystalline state. Ring opening and epimerization are facilitated by propitious water molecules that are observed in the structures. Determination of Fapy•dG mutagenicity in wild type and Pol β knockdown HEK 293T cells indicates that Pol β contributes to G → T transversions but also suppresses G → A transitions. Complementary kinetic studies have determined that Fapy•dG promotes mutagenesis by decreasing the catalytic efficiency of dCMP insertion opposite Fapy•dG, thus reducing polymerase fidelity. Kinetic studies have determined that dCMP incorporation opposite the β-anomer is ∼90 times faster than the α-anomer. This research identifies the importance of anomer dynamics, a feature unique to formamidopyrimidines, when considering the incorporation of nucleotides opposite Fapy•dG and potentially the repair of this structurally unusual lesion.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, and Department of Cancer Biology, University of Kansas Medical Center, Kansas City, Kansas 66160, United States.