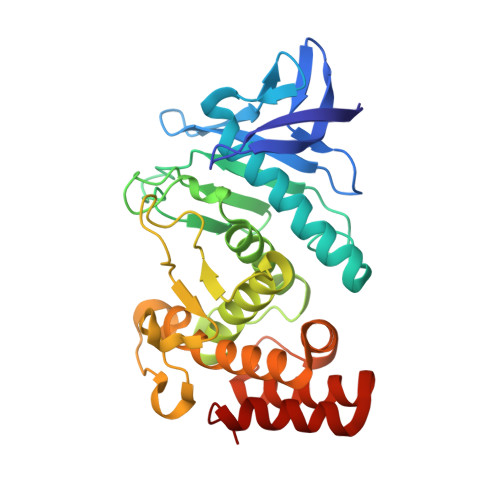
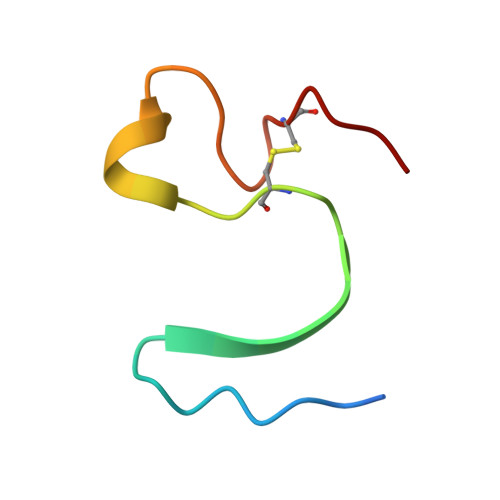
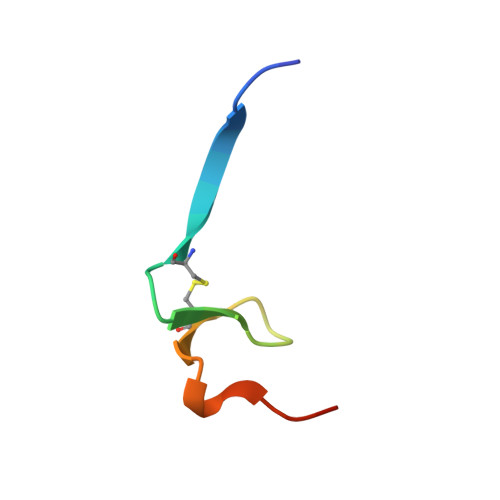
An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Mendes, S.R., Eckhard, U., Rodriguez-Banqueri, A., Guevara, T., Czermak, P., Marcos, E., Vilcinskas, A.(2022) Comput Struct Biotechnol J 20: 534-544