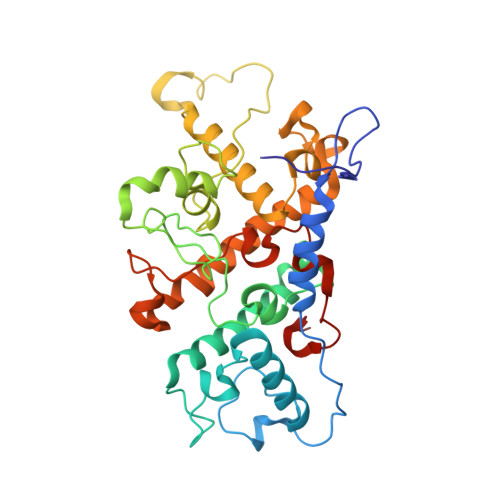
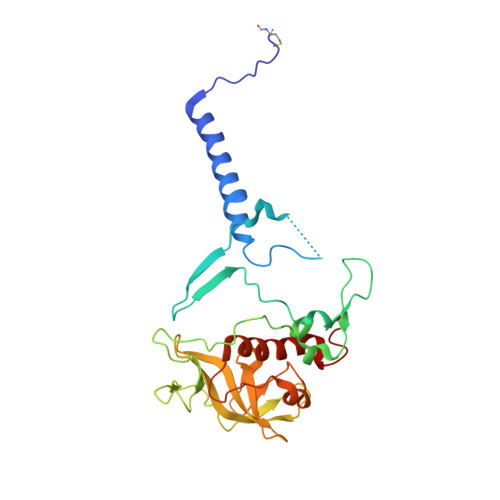
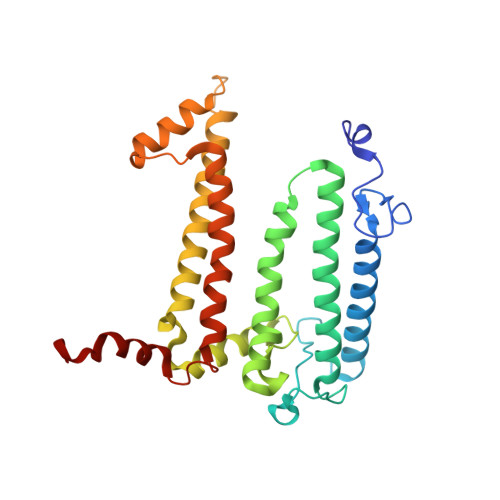
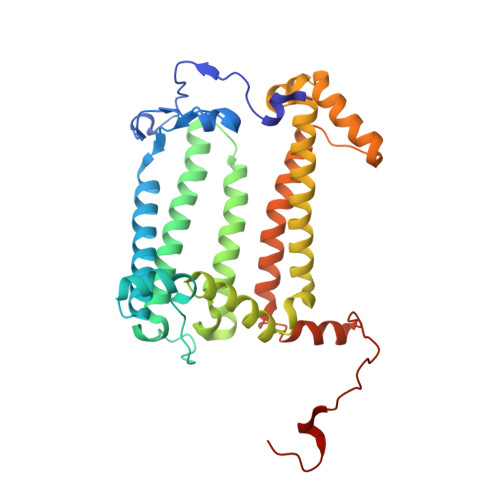
Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Bath, P., Banacore, A., Borjesson, P., Bosman, R., Wickstrand, C., Safari, C., Dods, R., Ghosh, S., Dahl, P., Ortolani, G., Bjorg Ulfarsdottir, T., Hammarin, G., Garcia Bonete, M.J., Vallejos, A., Ostojic, L., Edlund, P., Linse, J.B., Andersson, R., Nango, E., Owada, S., Tanaka, R., Tono, K., Joti, Y., Nureki, O., Luo, F., James, D., Nass, K., Johnson, P.J.M., Knopp, G., Ozerov, D., Cirelli, C., Milne, C., Iwata, S., Branden, G., Neutze, R.(2022) Acta Crystallogr D Struct Biol 78: 698-708
- PubMed: 35647917
- DOI: https://doi.org/10.1107/S2059798322004144
- Primary Citation of Related Structures:
7Q7P, 7Q7Q - PubMed Abstract:
Serial crystallography is a rapidly growing method that can yield structural insights from microcrystals that were previously considered to be too small to be useful in conventional X-ray crystallography. Here, conditions for growing microcrystals of the photosynthetic reaction centre of Blastochloris viridis within a lipidic cubic phase (LCP) crystallization matrix that employ a seeding protocol utilizing detergent-grown crystals with a different crystal packing are described. LCP microcrystals diffracted to 2.25 Å resolution when exposed to XFEL radiation, which is an improvement of 0.15 Å over previous microcrystal forms. Ubiquinone was incorporated into the LCP crystallization media and the resulting electron density within the mobile Q B pocket is comparable to that of other cofactors within the structure. As such, LCP microcrystallization conditions will facilitate time-resolved diffraction studies of electron-transfer reactions to the mobile quinone, potentially allowing the observation of structural changes associated with the two electron-transfer reactions leading to complete reduction of the ubiquinone ligand.
Organizational Affiliation:
Department of Chemistry and Molecular Biology, University of Gothenburg, Lundbergslaboratoriet Box 462, 405 30 Göteborg, Sweden.