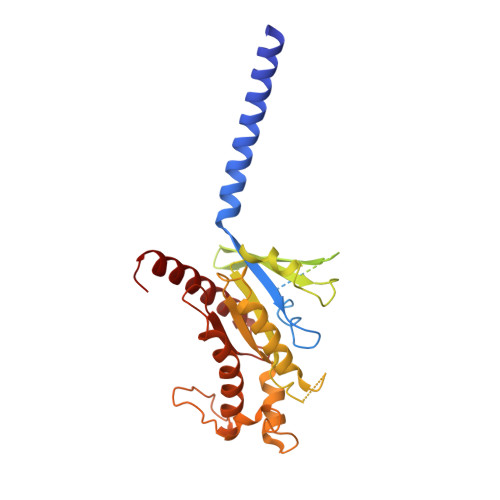
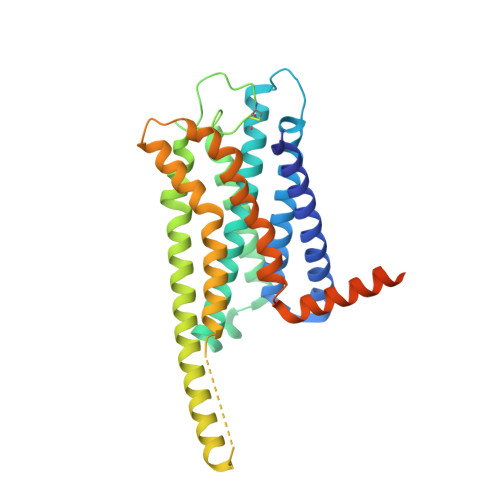
Activation and signaling mechanism revealed by GPR119-G s complex structures.
Qian, Y., Wang, J., Yang, L., Liu, Y., Wang, L., Liu, W., Lin, Y., Yang, H., Ma, L., Ye, S., Wu, S., Qiao, A.(2022) Nat Commun 13: 7033-7033
- PubMed: 36396650
- DOI: https://doi.org/10.1038/s41467-022-34696-6
- Primary Citation of Related Structures:
7WCM, 7WCN - PubMed Abstract:
Agonists selectively targeting cannabinoid receptor-like G-protein-coupled receptor (GPCR) GPR119 hold promise for treating metabolic disorders while avoiding unwanted side effects. Here we present the cryo-electron microscopy (cryo-EM) structures of the human GPR119-G s signaling complexes bound to AR231453 and MBX-2982, two representative agonists reported for GPR119. The structures reveal a one-amino acid shift of the conserved proline residue of TM5 that forms an outward bulge, opening up a hydrophobic cavity between TM4 and TM5 at the middle of the membrane for its endogenous ligands-monounsaturated lipid metabolites. In addition, we observed a salt bridge between ICL1 of GPR119 and Gβ s . Disruption of the salt bridge eliminates the cAMP production of GPR119, indicating an important role of Gβ s in GPR119-mediated signaling. Our structures, together with mutagenesis studies, illustrate the conserved binding mode of the chemically different agonists, and provide insights into the conformational changes in receptor activation and G protein coupling.
Organizational Affiliation:
Frontiers Science Center for Synthetic Biology (Ministry of Education), Tianjin Key Laboratory of Function and Application of Biological Macromolecular Structures, School of Life Sciences, Tianjin University, Tianjin, P. R. China.