Characterization of a novel cysteine-less Cu/Zn-superoxide dismutase in Paenibacillus lautus missing a conserved disulfide bond.
Furukawa, Y., Shintani, A., Narikiyo, S., Sue, K., Akutsu, M., Muraki, N.(2023) J Biological Chem 299: 105040-105040
- PubMed: 37442237
- DOI: https://doi.org/10.1016/j.jbc.2023.105040
- Primary Citation of Related Structures:
8IMD - PubMed Abstract:
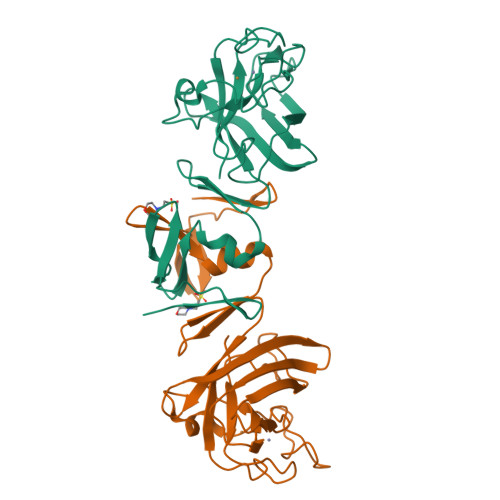
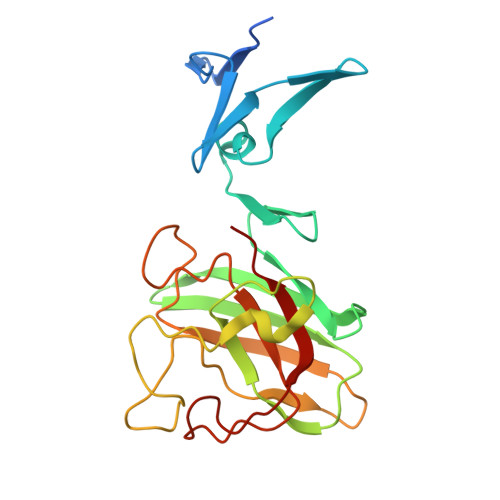
Cu/Zn-superoxide dismutase (CuZnSOD) is an enzyme that binds a copper and zinc ion and also forms an intramolecular disulfide bond. Together with the copper ion as the active site, the disulfide bond is completely conserved among these proteins; indeed, the disulfide bond plays critical roles in maintaining the catalytically competent conformation of CuZnSOD. Here, we found that a CuZnSOD protein in Paenibacillus lautus (PaSOD) has no Cys residue but exhibits a significant level of enzyme activity. The crystal structure of PaSOD revealed hydrophobic and hydrogen-bonding interactions in substitution for the disulfide bond of the other CuZnSOD proteins. Also notably, we determined that PaSOD forms a homodimer through an additional domain with a novel fold at the N terminus. While the advantages of lacking Cys residues and adopting a novel dimer configuration remain obscure, PaSOD does not require a disulfide-introducing/correcting system for maturation and could also avoid misfolding caused by aberrant thiol oxidations under an oxidative environment.
Organizational Affiliation:
Department of Chemistry, Keio University, Yokohama, Japan. Electronic address: furukawa@chem.keio.ac.jp.