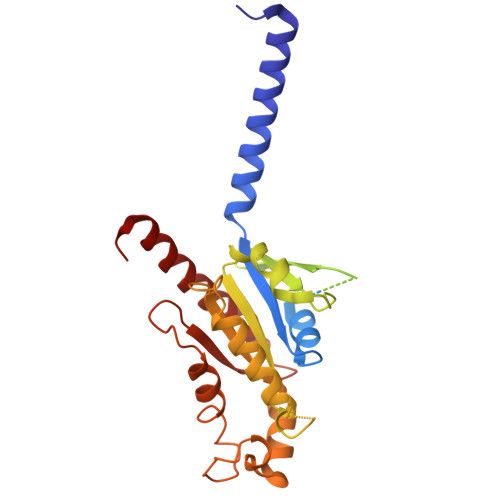
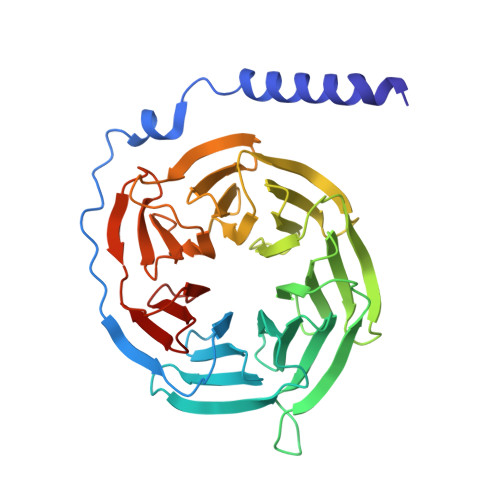
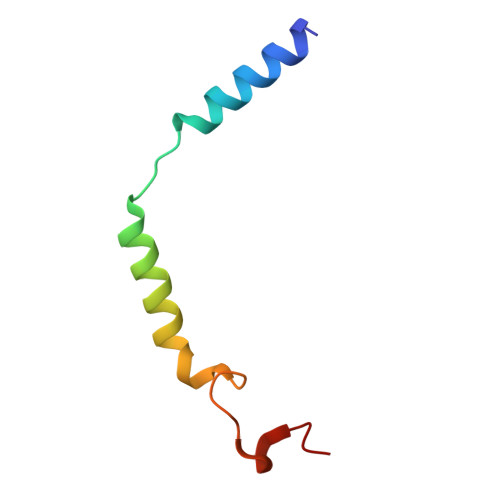
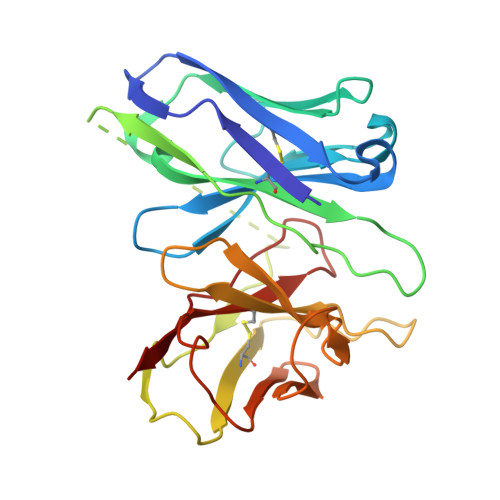
Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3.
Ye, F., Pan, X., Zhang, Z., Xiang, X., Li, X., Zhang, B., Ning, P., Liu, A., Wang, Q., Gong, K., Li, J., Zhu, L., Qian, C., Chen, G., Du, Y.(2024) Cell Rep 43: 114895-114895
- PubMed: 39427321
- DOI: https://doi.org/10.1016/j.celrep.2024.114895
- Primary Citation of Related Structures:
8JEI - PubMed Abstract:
Hydroxycarboxylic acid receptor 3 (HCAR3), a class A G-protein-coupled receptor, is an important cellular energy metabolism sensor with a key role in the regulation of lipolysis in humans. HCAR3 is deeply involved in many physiological processes and serves as a valuable target for the treatment of metabolic diseases, tumors, and immune diseases. Here, we report four cryoelectron microscopy (cryo-EM) structures of human HCAR3-Gi1 complexes with or without agonists: the endogenous ligand 3-hydroxyoctanoic acid, the drug niacin, the highly subtype-specific agonist compound 5c (4-(n-propyl)amino-3-nitrobenzoic acid), and the apo form. Together with mutagenesis and functional analyses, we revealed the recognition mechanisms of HCAR3 for different agonists. In addition, the key residues that determine the ligand selectivity between HCAR2 and HCAR3 were also illuminated. Overall, these findings provide a structural basis for the ligand recognition, activation, and selectivity and G-protein coupling mechanisms of HCAR3, which contribute to the design of HCAR3-targeting drugs with high efficacy and selectivity.
Organizational Affiliation:
Kobilka Institute of Innovative Drug Discovery, School of Medicine, the Chinese University of Hong Kong, Shenzhen 518172, Guangdong, China; Guangxi Key Laboratory of Special Biomedicine, School of Medicine, Guangxi University, Nanning 530004, China.