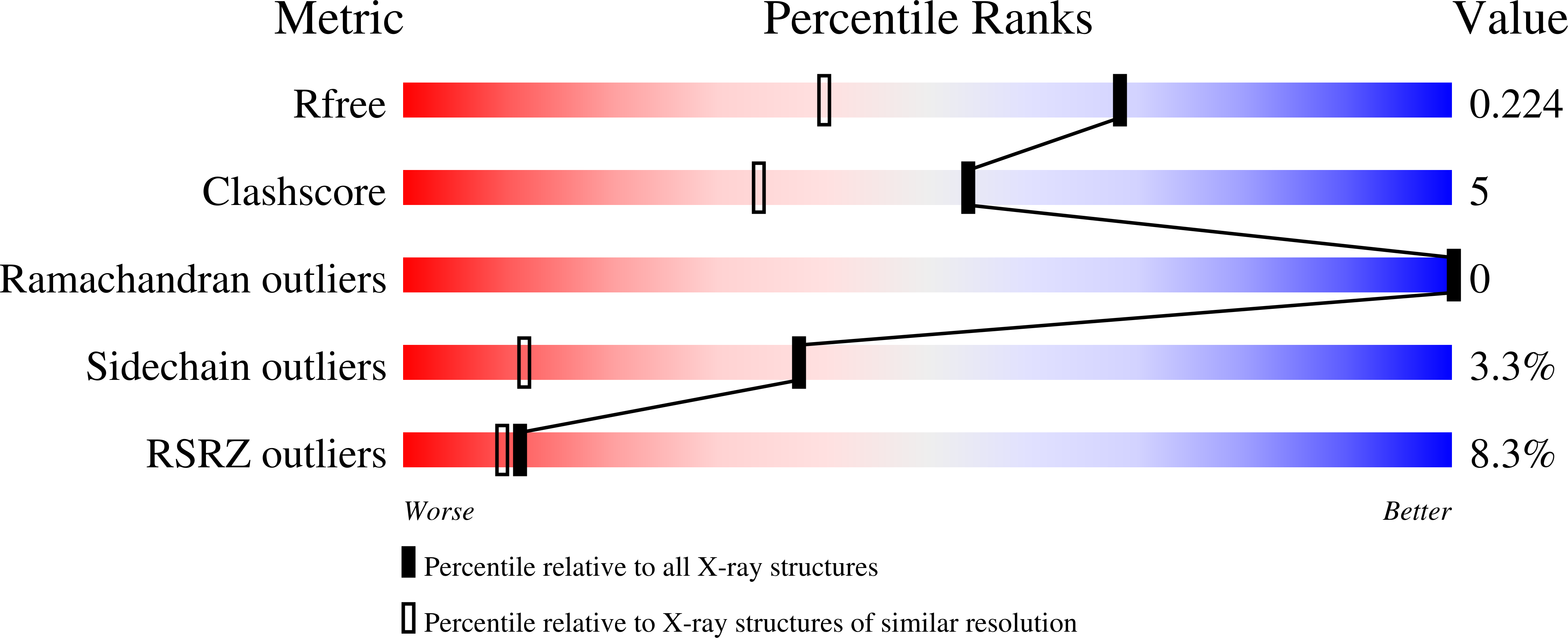
Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
De Rose, S.A., Isupov, M.N., Worthy, H.L., Stracke, C., Harmer, N.J., Siebers, B., Littlechild, J.A.(2023) Front Microbiol 14: 1267570-1267570
- PubMed: 38045033
- DOI: https://doi.org/10.3389/fmicb.2023.1267570
- Primary Citation of Related Structures:
8ORK, 8ORU - PubMed Abstract:
The enzyme cyclic di-phosphoglycerate synthetase that is involved in the production of the osmolyte cyclic 2,3-diphosphoglycerate has been studied both biochemically and structurally. Cyclic 2,3-diphosphoglycerate is found exclusively in the hyperthermophilic archaeal methanogens, such as Methanothermus fervidus , Methanopyrus kandleri , and Methanothermobacter thermoautotrophicus . Its presence increases the thermostability of archaeal proteins and protects the DNA against oxidative damage caused by hydroxyl radicals. The cyclic 2,3-diphosphoglycerate synthetase enzyme has been crystallized and its structure solved to 1.7 Å resolution by experimental phasing. It has also been crystallized in complex with its substrate 2,3 diphosphoglycerate and the co-factor ADP and this structure has been solved to 2.2 Å resolution. The enzyme structure has two domains, the core domain shares some structural similarity with other NTP-dependent enzymes. A significant proportion of the structure, including a 127 amino acid N-terminal domain, has no structural similarity to other known enzyme structures. The structure of the complex shows a large conformational change that occurs in the enzyme during catalytic turnover. The reaction involves the transfer of the γ-phosphate group from ATP to the substrate 2,3 -diphosphoglycerate and the subsequent S N 2 attack to form a phosphoanhydride. This results in the production of the unusual extremolyte cyclic 2,3 -diphosphoglycerate which has important industrial applications.
Organizational Affiliation:
Henry Wellcome Building for Biocatalysis, Biosciences, Faculty of Health and Life Sciences, University of Exeter, Exeter, United Kingdom.