Human 2'-Deoxynucleoside 5'-Phosphate N -Hydrolase 1: Mechanism of 2'-Deoxyuridine 5'-Monophosphate Hydrolysis.
Devi, S., Carberry, A.E., Zickuhr, G.M., Dickson, A.L., Harrison, D.J., da Silva, R.G.(2023) Biochemistry 62: 2658-2668
- PubMed: 37582341
- DOI: https://doi.org/10.1021/acs.biochem.3c00369
- Primary Citation of Related Structures:
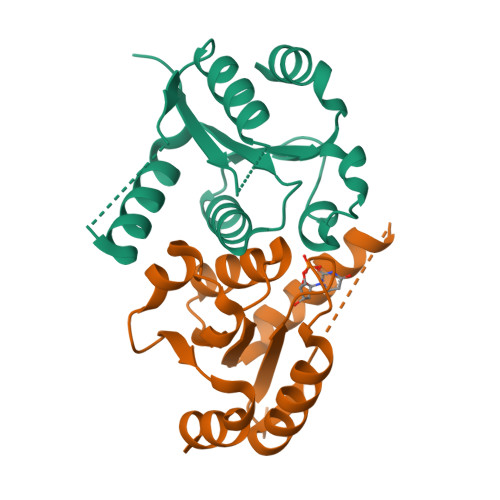
8OS9, 8OSC - PubMed Abstract:
The enzyme 2'-deoxynucleoside 5'-phosphate N -hydrolase 1 (DNPH1) catalyzes the N -ribosidic bond cleavage of 5-hydroxymethyl-2'-deoxyuridine 5'-monophosphate to generate 2-deoxyribose 5-phosphate and 5-hydroxymethyluracil. DNPH1 accepts other 2'-deoxynucleoside 5'-monophosphates as slow-reacting substrates. DNPH1 inhibition is a promising strategy to overcome resistance to and potentiate anticancer poly(ADP-ribose) polymerase inhibitors. We solved the crystal structure of unliganded human DNPH1 and took advantage of the slow reactivity of 2'-deoxyuridine 5'-monophosphate (dUMP) as a substrate to obtain a crystal structure of the DNPH1:dUMP Michaelis complex. In both structures, the carboxylate group of the catalytic Glu residue, proposed to act as a nucleophile in covalent catalysis, forms an apparent low-barrier hydrogen bond with the hydroxyl group of a conserved Tyr residue. The crystal structures are supported by functional data, with liquid chromatography-mass spectrometry analysis showing that DNPH1 incubation with dUMP leads to slow yet complete hydrolysis of the substrate. A direct UV-vis absorbance-based assay allowed characterization of DNPH1 kinetics at low dUMP concentrations. A bell-shaped pH-rate profile indicated that acid-base catalysis is operational and that for maximum k cat / K M , two groups with an average p K a of 6.4 must be deprotonated, while two groups with an average p K a of 8.2 must be protonated. A modestly inverse solvent viscosity effect rules out diffusional processes involved in dUMP binding to and possibly uracil release from the enzyme as rate limiting to k cat / K M . Solvent deuterium isotope effects on k cat / K M and k cat were inverse and unity, respectively. A reaction mechanism for dUMP hydrolysis is proposed.
Organizational Affiliation:
School of Biology, Biomedical Sciences Research Complex, University of St Andrews, St Andrews KY16 9ST, U.K.