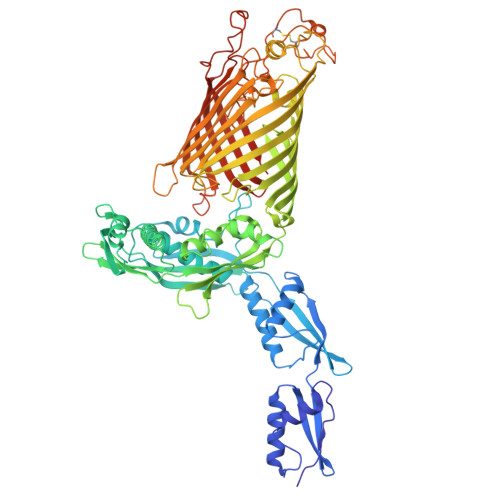
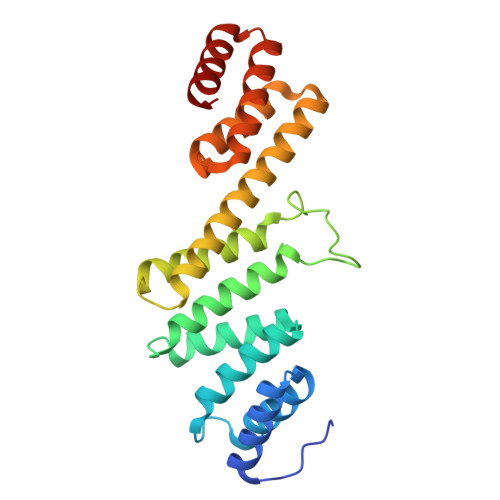
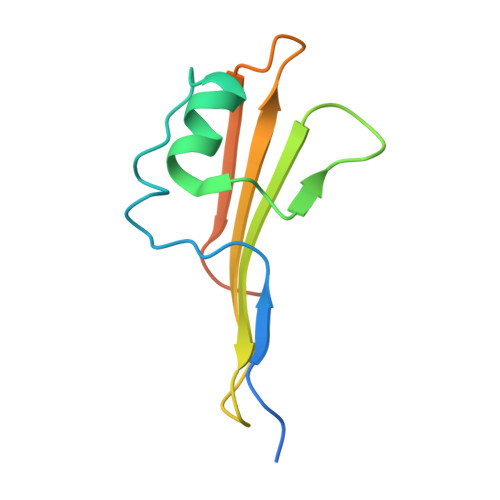
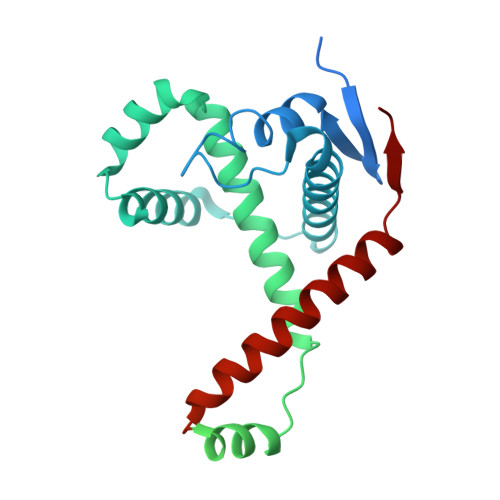
Outer membrane protein assembly mediated by BAM-SurA complexes.
Fenn, K.L., Horne, J.E., Crossley, J.A., Bohringer, N., Horne, R.J., Schaberle, T.F., Calabrese, A.N., Radford, S.E., Ranson, N.A.(2024) Nat Commun 15: 7612-7612
- PubMed: 39218969
- DOI: https://doi.org/10.1038/s41467-024-51358-x
- Primary Citation of Related Structures:
8PZ1, 8PZ2, 8PZU, 8PZV, 8Q0G, 8QP5, 8QPU, 8QPV, 8QPW - PubMed Abstract:
The outer membrane is a formidable barrier that protects Gram-negative bacteria against environmental threats. Its integrity requires the correct folding and insertion of outer membrane proteins (OMPs) by the membrane-embedded β-barrel assembly machinery (BAM). Unfolded OMPs are delivered to BAM by the periplasmic chaperone SurA, but how SurA and BAM work together to ensure successful OMP delivery and folding remains unclear. Here, guided by AlphaFold2 models, we use disulphide bond engineering in an attempt to trap SurA in the act of OMP delivery to BAM, and solve cryoEM structures of a series of complexes. The results suggest that SurA binds BAM at its soluble POTRA-1 domain, which may trigger conformational changes in both BAM and SurA that enable transfer of the unfolded OMP to the BAM lateral gate for insertion into the outer membrane. Mutations that disrupt the interaction between BAM and SurA result in outer membrane assembly defects, supporting the key role of SurA in outer membrane biogenesis.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology and School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, LS2 9JT, UK.