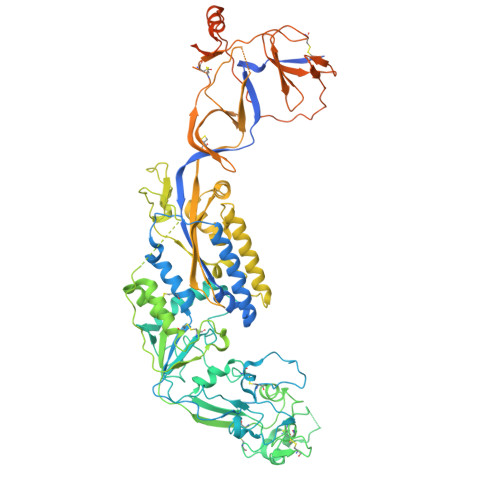
Structures of the Foamy virus fusion protein reveal an unexpected link with the F protein of paramyxo- and pneumoviruses.
Fernandez, I., Bontems, F., Brun, D., Coquin, Y., Goverde, C.A., Correia, B.E., Gessain, A., Buseyne, F., Rey, F.A., Backovic, M.(2024) Sci Adv 10: eado7035-eado7035
- PubMed: 39392890
- DOI: https://doi.org/10.1126/sciadv.ado7035
- Primary Citation of Related Structures:
8RM0, 8RM1 - PubMed Abstract:
Foamy viruses (FVs) constitute a subfamily of retroviruses. Their envelope (Env) glycoprotein drives the merger of viral and cellular membranes during entry into cells. The only available structures of retroviral Envs are those from human and simian immunodeficiency viruses from the subfamily of orthoretroviruses, which are only distantly related to the FVs. We report the cryo-electron microscopy structures of the FV Env ectodomain in the pre- and post-fusion states, which unexpectedly demonstrate structural similarity with the fusion protein (F) of paramyxo- and pneumoviruses, implying an evolutionary link between the viral fusogens. We describe the structural features that are unique to the FV Env and propose a mechanistic model for its conformational change, highlighting how the interplay of its structural elements could drive membrane fusion and viral entry. The structural knowledge on the FV Env now provides a framework for functional investigations, which can benefit the design of FV Env variants with improved features for use as gene therapy vectors.
- Institut Pasteur, Université Paris Cité, CNRS UMR3569, Unité de Virologie Structurale, 75015 Paris, France.
Organizational Affiliation: