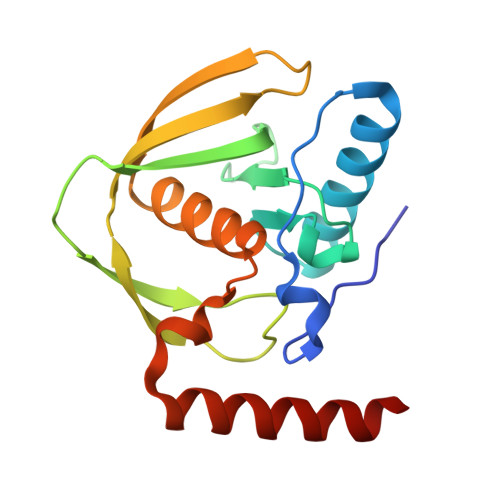
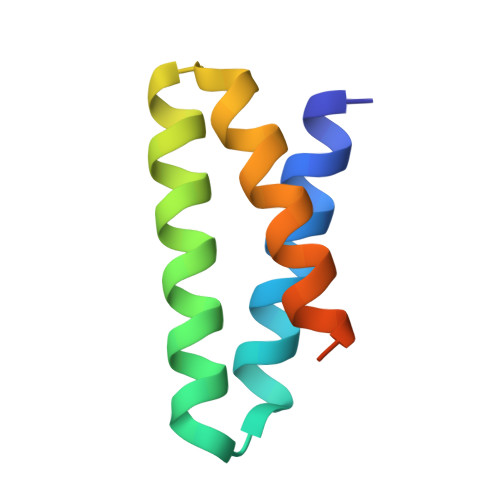
Targeting hybrid neosurface fingerprints for the design of de novo drug-induced protein interactions
Marchand, A., Buckley, S., Schneuing, A., Pacesa, M., Gainza, P., Elizarova, E., Neeser, R., Reymond, L., Georgeon, S., Schmidt, J., Correia, B.E.To be published.