Cryo-EM complex of DDM1-HELLS chimera bound to the nucleosome
Nartey, W., Williams, G.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent DNA helicase DDM1,Lymphoid-specific helicase chimera | 821 | Arabidopsis thaliana, Homo sapiens | Mutation(s): 0 Gene Names: DDM1, HELLS EC: 3.6.4 (UniProt), 3.6.4.12 (UniProt) | 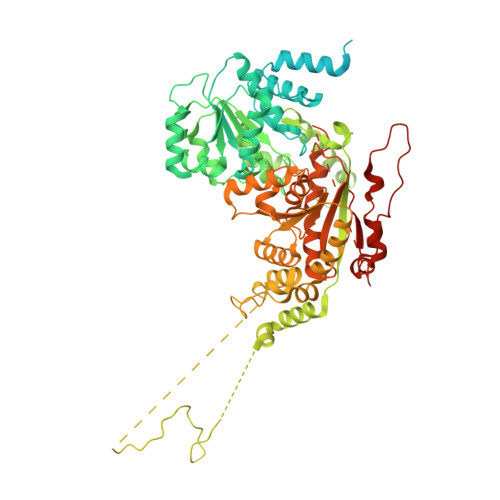 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9XFH4 (Arabidopsis thaliana) Explore Q9XFH4 Go to UniProtKB: Q9XFH4 | |||||
Find proteins for Q9NRZ9 (Homo sapiens) Explore Q9NRZ9 Go to UniProtKB: Q9NRZ9 | |||||
PHAROS: Q9NRZ9 GTEx: ENSG00000119969 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q9NRZ9Q9XFH4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H3.2 | 136 | Xenopus laevis | Mutation(s): 1 | 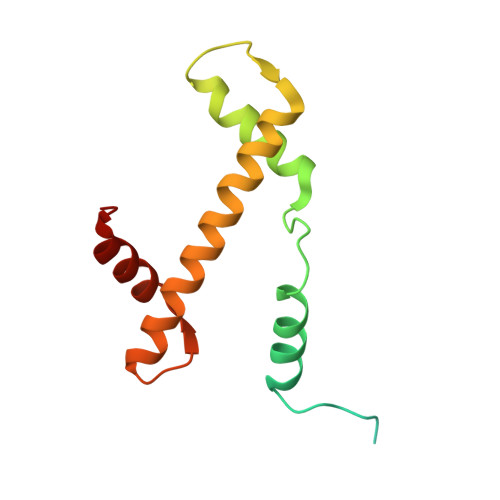 | |
UniProt | |||||
Find proteins for P84233 (Xenopus laevis) Explore P84233 Go to UniProtKB: P84233 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P84233 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A | 133 | Xenopus laevis | Mutation(s): 0 Gene Names: h2ac14.L | 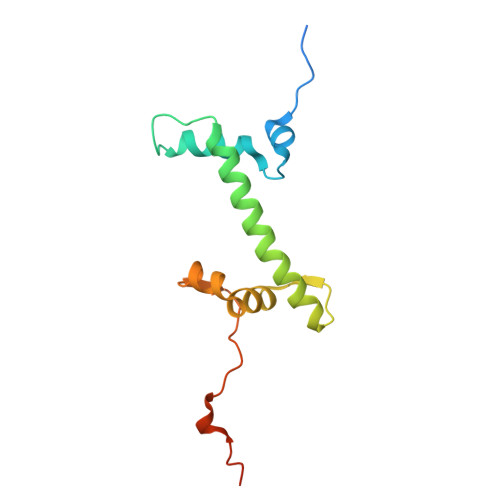 | |
UniProt | |||||
Find proteins for Q6AZJ8 (Xenopus laevis) Explore Q6AZJ8 Go to UniProtKB: Q6AZJ8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6AZJ8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2B 1.1 | 123 | Xenopus laevis | Mutation(s): 1 | 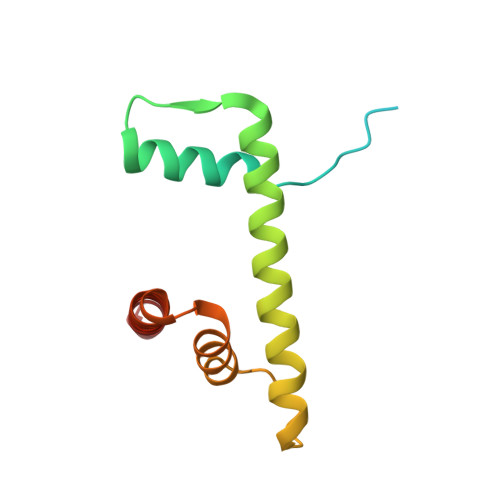 | |
UniProt | |||||
Find proteins for P02281 (Xenopus laevis) Explore P02281 Go to UniProtKB: P02281 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02281 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H4 | 110 | Xenopus laevis | Mutation(s): 0 Gene Names: LOC121398084 | 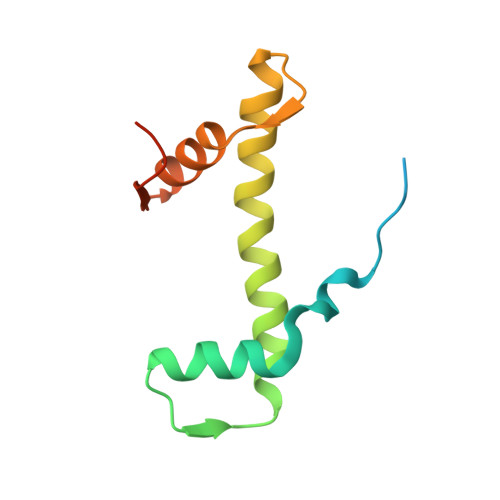 | |
UniProt | |||||
Find proteins for A0A8J1LTD2 (Xenopus laevis) Explore A0A8J1LTD2 Go to UniProtKB: A0A8J1LTD2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8J1LTD2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (192-MER) | 192 | Homo sapiens | 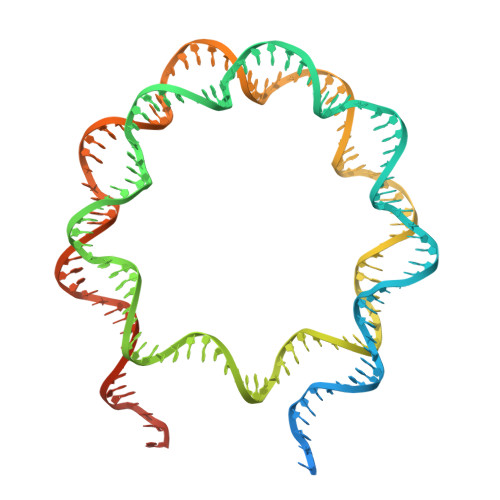 | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (192-MER) | 192 | Homo sapiens | 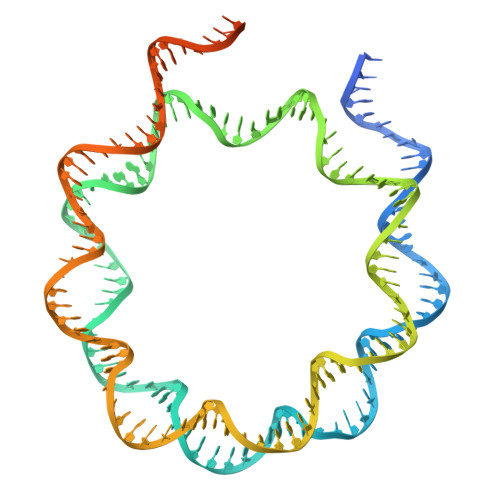 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | L [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| BEF (Subject of Investigation/LOI) Query on BEF | M [auth A] | BERYLLIUM TRIFLUORIDE ION Be F3 OGIAHMCCNXDTIE-UHFFFAOYSA-K |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 3.3.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | RGPIN-2018-04327 |