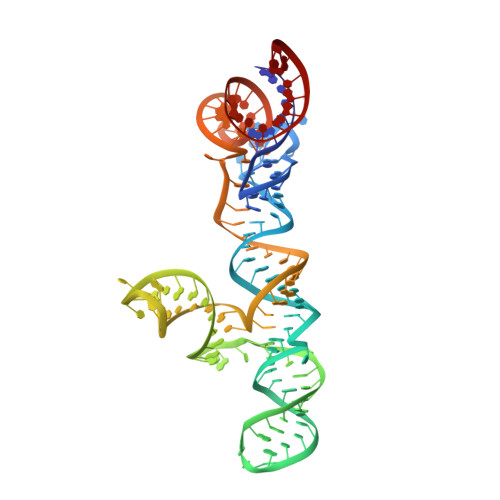
Structure of HIV-1 RRE stem-loop II identifies two conformational states of the high-affinity Rev binding site.
Tipo, J., Gottipati, K., Slaton, M., Gonzalez-Gutierrez, G., Choi, K.H.(2024) Nat Commun 15: 4198-4198
- PubMed: 38760344
- DOI: https://doi.org/10.1038/s41467-024-48162-y
- Primary Citation of Related Structures:
8UO6 - PubMed Abstract:
During HIV infection, specific RNA-protein interaction between the Rev response element (RRE) and viral Rev protein is required for nuclear export of intron-containing viral mRNA transcripts. Rev initially binds the high-affinity site in stem-loop II, which promotes oligomerization of additional Rev proteins on RRE. Here, we present the crystal structure of RRE stem-loop II in distinct closed and open conformations. The high-affinity Rev-binding site is located within the three-way junction rather than the predicted stem IIB. The closed and open conformers differ in their non-canonical interactions within the three-way junction, and only the open conformation has the widened major groove conducive to initial Rev interaction. Rev binding assays show that RRE stem-loop II has high- and low-affinity binding sites, each of which binds a Rev dimer. We propose a binding model, wherein Rev-binding sites on RRE are sequentially created through structural rearrangements induced by Rev-RRE interactions.
- Department of Pharmacology and Toxicology, The University of Texas Medical Branch, Galveston, TX, 77555, USA.
Organizational Affiliation: