Differentiating MHC-II antigens with a single loop
Du, H., Liu, J., Jude, K.M., Yang, X., Li, Y., Bell, B., Yang, H., Kassardjian, A., Mobedi, A., Parekh, U., Sperberg, A., Julien, J.-P., Mellins, E., Garcia, K.C., Huang, P.-S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HLA class II histocompatibility antigen, DR alpha chain | 184 | Homo sapiens | Mutation(s): 0 Gene Names: HLA-DRA, HLA-DRA1 | 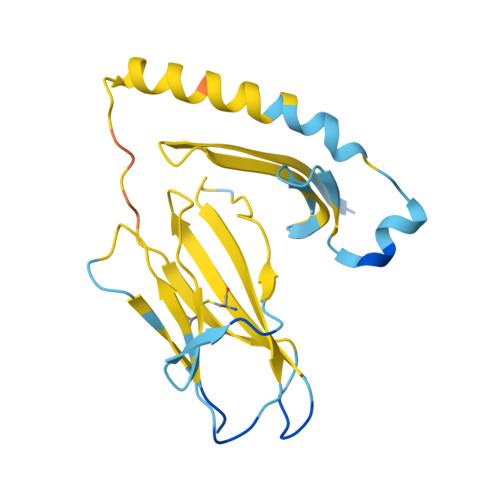 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01903 (Homo sapiens) Explore P01903 Go to UniProtKB: P01903 | |||||
PHAROS: P01903 GTEx: ENSG00000204287 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01903 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P01903-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HLA class II histocompatibility antigen DR beta chain | 192 | Homo sapiens | Mutation(s): 0 Gene Names: HLA-DRB1 | 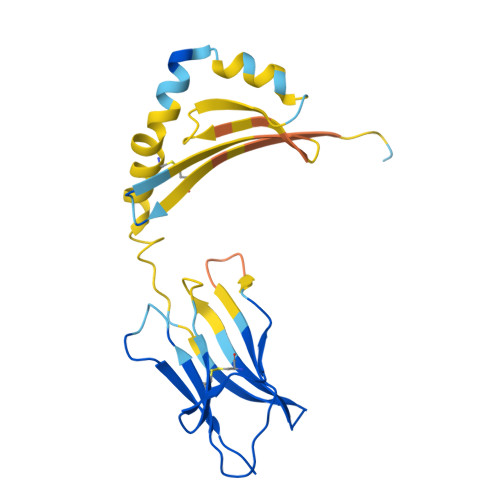 | |
UniProt | |||||
Find proteins for D7RIG0 (Homo sapiens) Explore D7RIG0 Go to UniProtKB: D7RIG0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D7RIG0 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Superantigen | 129 | Metamycoplasma arthritidis | Mutation(s): 18 | 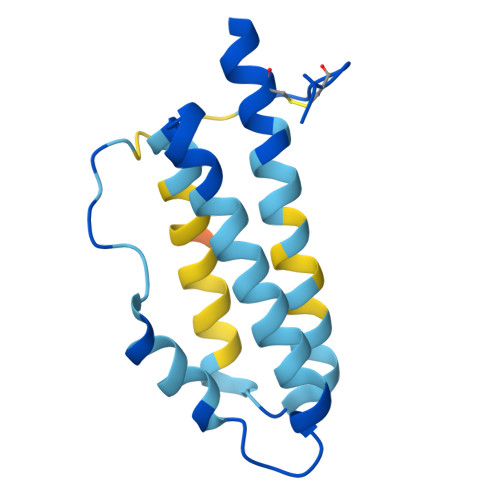 | |
UniProt | |||||
Find proteins for Q48898 (Metamycoplasma arthritidis) Explore Q48898 Go to UniProtKB: Q48898 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q48898 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| c44H10 Fab heavy chain | D [auth H] | 223 | Homo sapiens | Mutation(s): 0 | 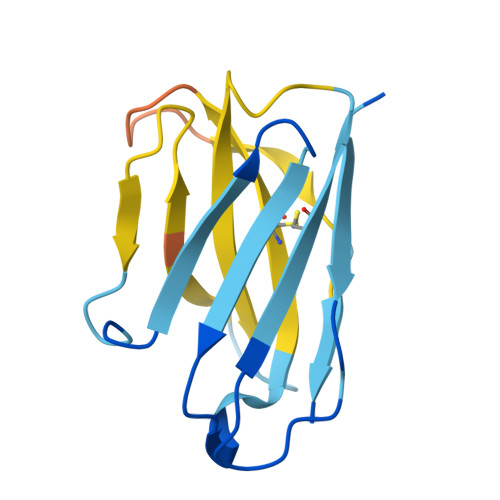 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| c44H10 Fab light chain | E [auth L] | 214 | Homo sapiens | Mutation(s): 0 | 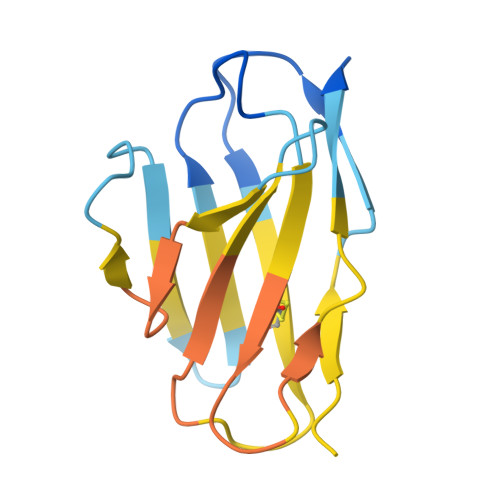 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Class-II-associated invariant chain peptide | F [auth P] | 15 | Homo sapiens | Mutation(s): 0 Gene Names: CD74, DHLAG |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04233 (Homo sapiens) Explore P04233 Go to UniProtKB: P04233 | |||||
PHAROS: P04233 GTEx: ENSG00000019582 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04233 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | G [auth A], H [auth A], I [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21_5207 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | r01gm147893 |