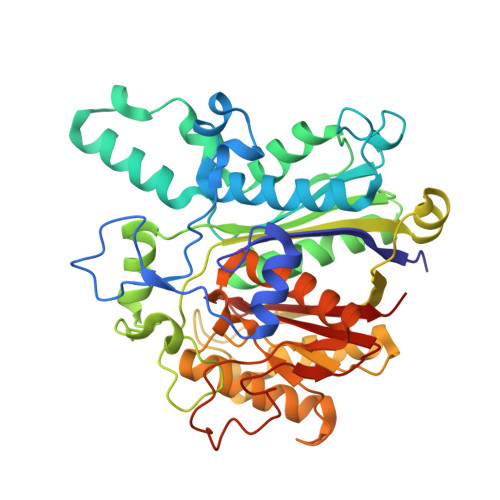
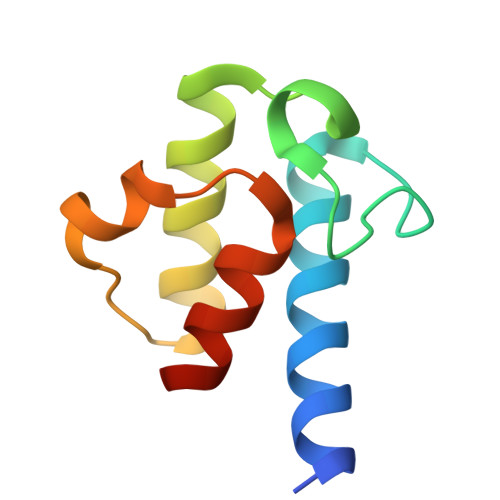
The beta-Ketoacyl-ACP Synthase FabF Catalyzes Carbon-Carbon Bond Formation in a Bimodal Pattern for Fatty Acid Biosynthesis.
Huang, Y., Wang, Y., Cai, C., Zhang, L., Ye, F., Zhang, L.(2024) Angew Chem Int Ed Engl 63: e202407921-e202407921
- PubMed: 39175097
- DOI: https://doi.org/10.1002/anie.202407921
- Primary Citation of Related Structures:
8Z5C, 8Z5D, 8Z5E, 8Z5F - PubMed Abstract:
Fatty acids produced by the type-II fatty acid biosynthesis pathway (FAS-II) are essential biomaterials for bacterial membrane construction and numerous metabolic routes. The β-ketoacyl-ACP synthase FabF catalyzes the key C-C bond formation step for fatty acid extension in FAS-II. Here, we revealed the substrate recognition and catalytic mechanisms of FabF by determining FabF-ACP complexes. FabF displays a distinctive bimodal catalytic pattern specifically on C6 and C10 acyl-ACP substrates. It utilizes positively charged residues located on the η3-helix and loop1 regions near the catalytic tunnel entrance to bind ACP, and two hydrophobic cavities as well as "front", "middle", and "back" door residues to specifically stabilize C6 and C10 acyl substrates for preferential catalysis. Further quantum chemistry calculations suggest that the FabF catalytic residues Lys336 and His304 facilitate proton transfer during condensation catalysis and C-C bond formation. Our results provide key mechanistic insights into the biosynthesis of molecular carbon skeletons based on ketosynthases that are highly conserved through the FAS and polyketide synthase (PKS) analogous biosynthetic routes, broaden the understanding of the tricarboxylic acid cycle that utilizes lipoic acid derived from C8-ACP accumulated due to the FabF distinctive catalytic pattern for oxidative decarboxylations, and may facilitate the development of narrow-spectrum antibacterial drugs.
Organizational Affiliation:
Shanghai Jiao Tong University School of Medicine, Pharmacology and chemical biology, Building #3, Room #213,, No.280 South Chongqing Road, 200025, Shanghai, CHINA.