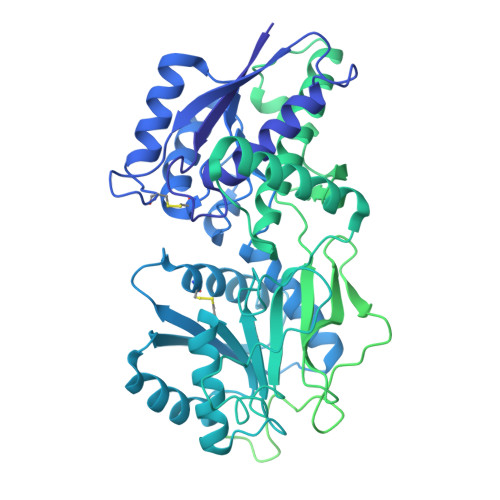
Architecture and activation of single-pass transmembrane receptor guanylyl cyclase.
Liu, S., Payne, A.M., Wang, J., Zhu, L., Paknejad, N., Eng, E.T., Liu, W., Miao, Y., Hite, R.K., Huang, X.Y.(2025) Nat Struct Mol Biol 32: 469-478
- PubMed: 39543315
- DOI: https://doi.org/10.1038/s41594-024-01426-z
- Primary Citation of Related Structures:
9BCL, 9BCN, 9BCO, 9BCP, 9BCQ, 9BCS, 9BCV - PubMed Abstract:
The heart, in addition to its primary role in blood circulation, functions as an endocrine organ by producing cardiac hormone natriuretic peptides. These hormones regulate blood pressure through the single-pass transmembrane receptor guanylyl cyclase A (GC-A), also known as natriuretic peptide receptor 1. The binding of the peptide hormones to the extracellular domain of the receptor activates the intracellular guanylyl cyclase domain of the receptor to produce the second messenger cyclic guanosine monophosphate. Despite their importance, the detailed architecture and domain interactions within full-length GC-A remain elusive. Here we present cryo-electron microscopy structures, functional analyses and molecular dynamics simulations of full-length human GC-A, in both the absence and the presence of atrial natriuretic peptide. The data reveal the architecture of full-length GC-A, highlighting the spatial arrangement of its various functional domains. This insight is crucial for understanding how different parts of the receptor interact and coordinate during activation. The study elucidates the molecular basis of how extracellular signals are transduced across the membrane to activate the intracellular guanylyl cyclase domain.
- Department of Physiology and Biophysics, Weill Cornell Medical College of Cornell University, New York, NY, USA.
Organizational Affiliation: