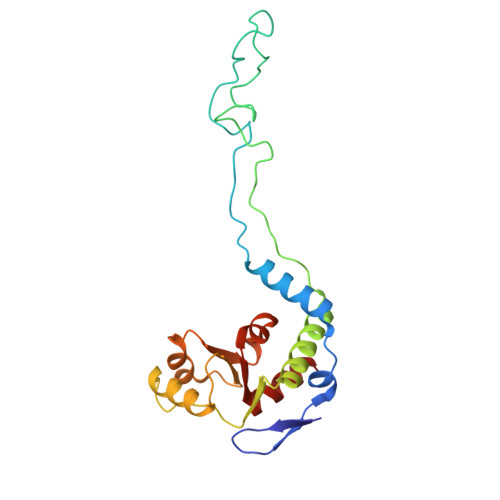
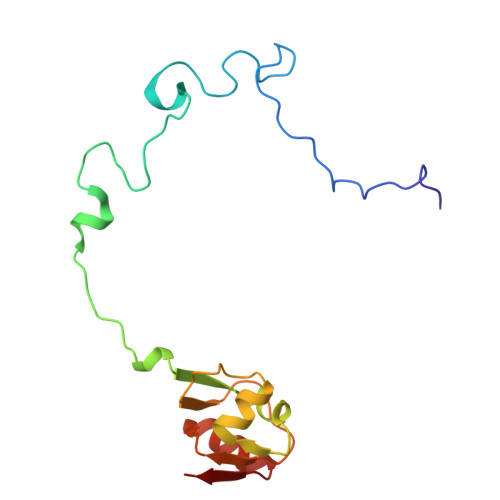
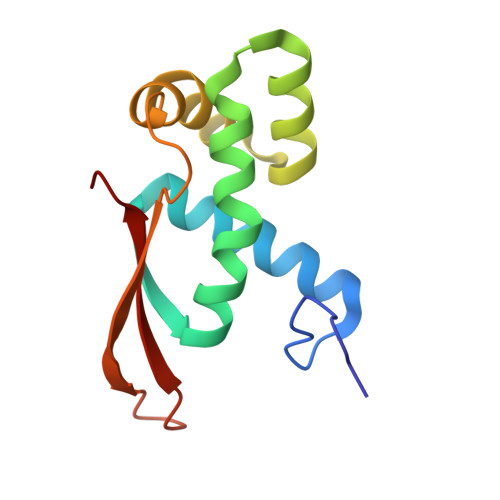
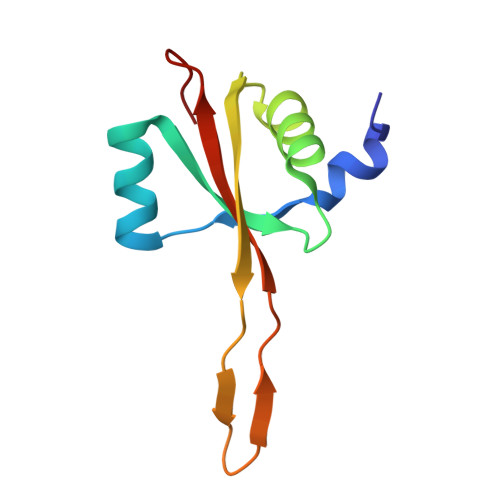
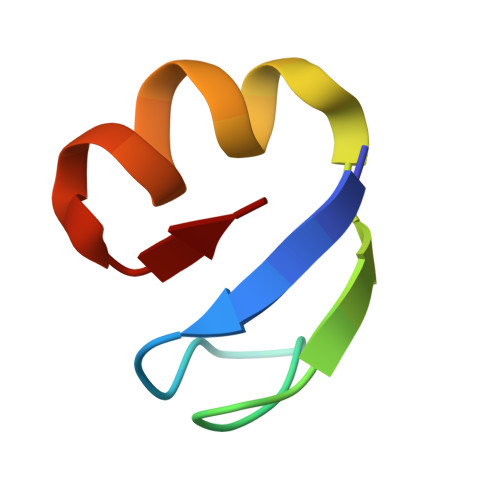
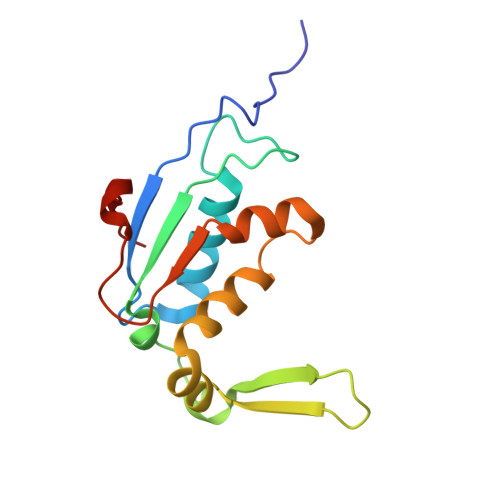
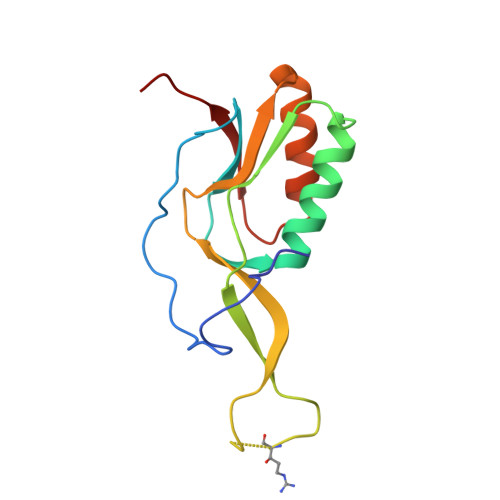
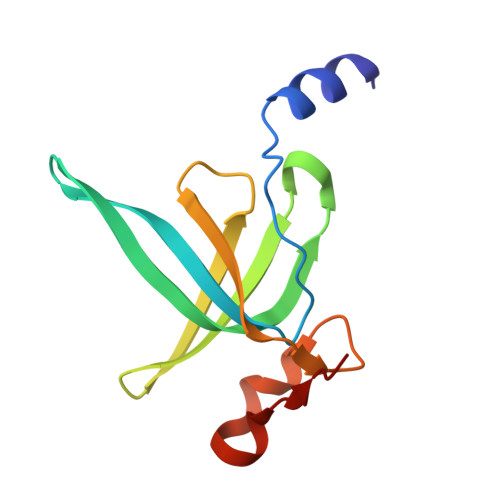
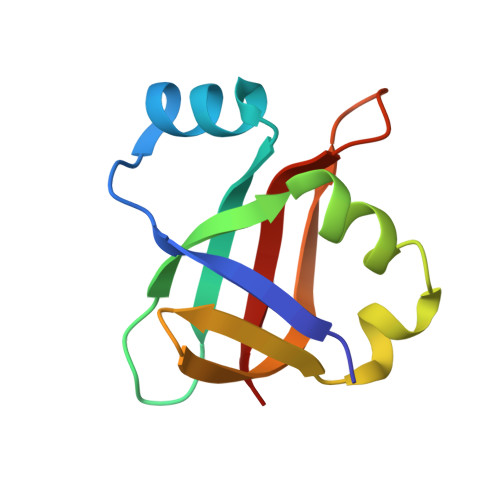
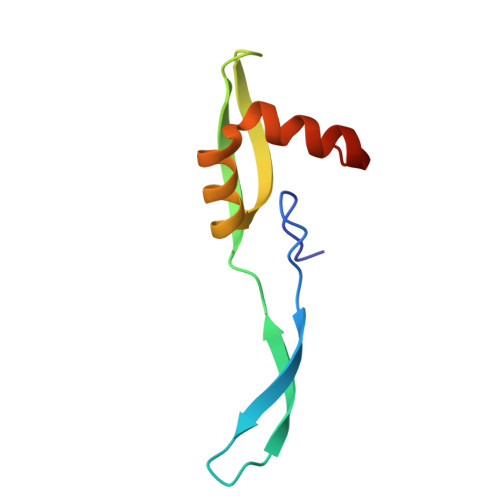
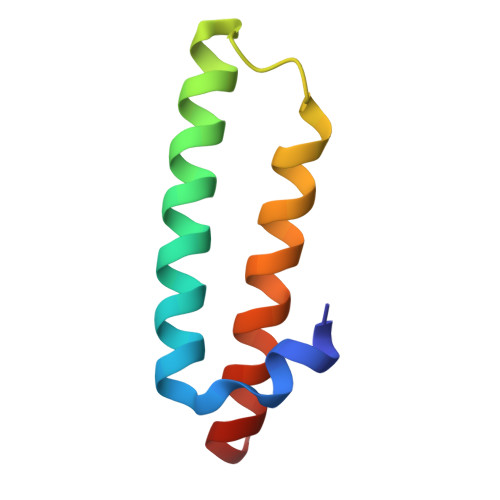
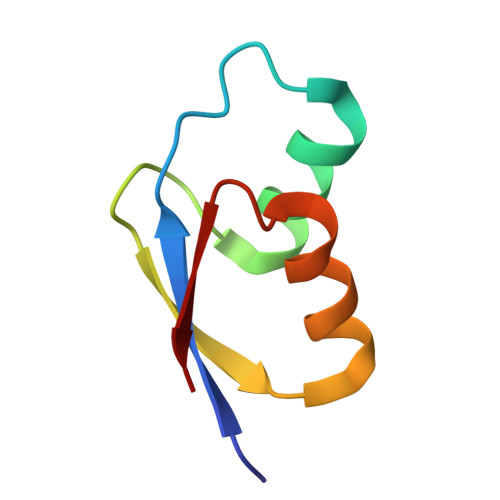
Rumicidins are a family of mammalian host-defense peptides plugging the 70S ribosome exit tunnel.
Panteleev, P.V., Pichkur, E.B., Kruglikov, R.N., Paleskava, A., Shulenina, O.V., Bolosov, I.A., Bogdanov, I.V., Safronova, V.N., Balandin, S.V., Marina, V.I., Kombarova, T.I., Korobova, O.V., Shamova, O.V., Myasnikov, A.G., Borzilov, A.I., Osterman, I.A., Sergiev, P.V., Bogdanov, A.A., Dontsova, O.A., Konevega, A.L., Ovchinnikova, T.V.(2024) Nat Commun 15: 8925-8925
- PubMed: 39414793
- DOI: https://doi.org/10.1038/s41467-024-53309-y
- Primary Citation of Related Structures:
9D89 - PubMed Abstract:
The antimicrobial resistance crisis along with challenges of antimicrobial discovery revealed the vital necessity to develop new antibiotics. Many of the animal proline-rich antimicrobial peptides (PrAMPs) inhibit the process of bacterial translation. Genome projects allowed to identify immune-related genes encoding animal host defense peptides. Here, using genome mining approach, we discovered a family of proline-rich cathelicidins, named rumicidins. The genes encoding these peptides are widespread among ruminant mammals. Biochemical studies indicated that rumicidins effectively inhibited the elongation stage of bacterial translation. The cryo-EM structure of the Escherichia coli 70S ribosome in complex with one of the representatives of the family revealed that the binding site of rumicidins span the ribosomal A-site cleft and the nascent peptide exit tunnel interacting with its constriction point by the conservative Trp23-Phe24 dyad. Bacterial resistance to rumicidins is mediated by knockout of the SbmA transporter or modification of the MacAB-TolC efflux pump. A wide spectrum of antibacterial activity, a high efficacy in the animal infection model, and lack of adverse effects towards human cells in vitro make rumicidins promising molecular scaffolds for development of ribosome-targeting antibiotics.
- M.M. Shemyakin & Yu.A. Ovchinnikov Institute of Bioorganic Chemistry, the Russian Academy of Sciences, Moscow, Russia.
Organizational Affiliation: