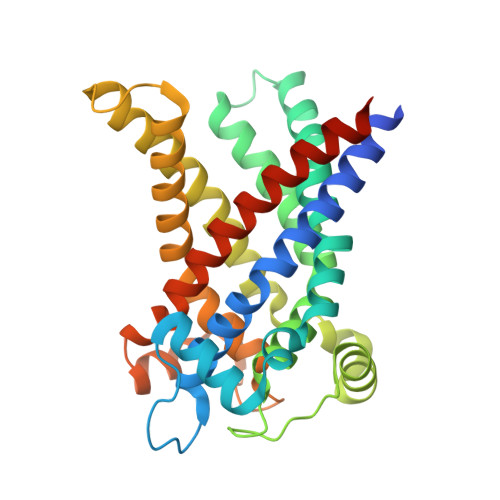
Proton conductance by human uncoupling protein 1 is inhibited by purine and pyrimidine nucleotides.
Jones, S.A., Sowton, A.P., Lacabanne, D., King, M.S., Palmer, S.M., Zogg, T., Pardon, E., Steyaert, J., Ruprecht, J.J., Kunji, E.R.S.(2025) EMBO J 44: 2353-2365
- PubMed: 40021843
- DOI: https://doi.org/10.1038/s44318-025-00395-3
- Primary Citation of Related Structures:
9FZQ - PubMed Abstract:
Uncoupling protein 1 (UCP1, SLC25A7) is responsible for the thermogenic properties of brown adipose tissue. Upon fatty acid activation, UCP1 facilitates proton leakage, dissipating the mitochondrial proton motive force to release energy as heat. Purine nucleotides are considered to be the only inhibitors of UCP1 activity, binding to its central cavity to lock UCP1 in a proton-impermeable conformation. Here we show that pyrimidine nucleotides can also bind and inhibit its proton-conducting activity. All nucleotides bound in a pH-dependent manner, with the highest binding affinity observed for ATP, followed by dTTP, UTP, GTP and CTP. We also determined the structural basis of UTP binding to UCP1, showing that binding of purine and pyrimidine nucleotides follows the same molecular principles. We find that the closely related mitochondrial dicarboxylate carrier (SLC25A10) and oxoglutarate carrier (SLC25A11) have many cavity residues in common, but do not bind nucleotides. Thus, while UCP1 has evolved from dicarboxylate carriers, no selection for nucleobase specificity has occurred, highlighting the importance of the pH-dependent nucleotide binding mechanism mediated via the phosphate moieties.
- MRC Mitochondrial Biology Unit, University of Cambridge, Cambridge Biomedical Campus, Keith Peters Building, Cambridge, CB2 0XY, UK.
Organizational Affiliation: