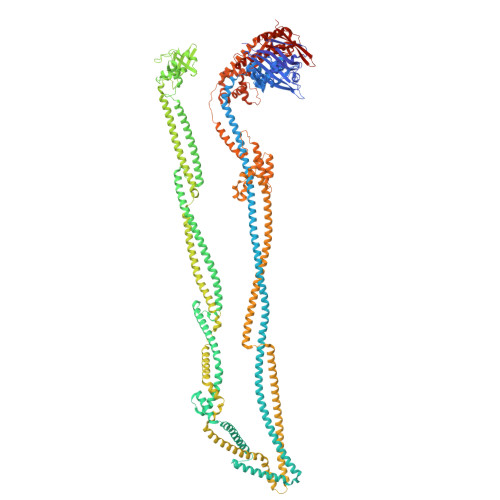
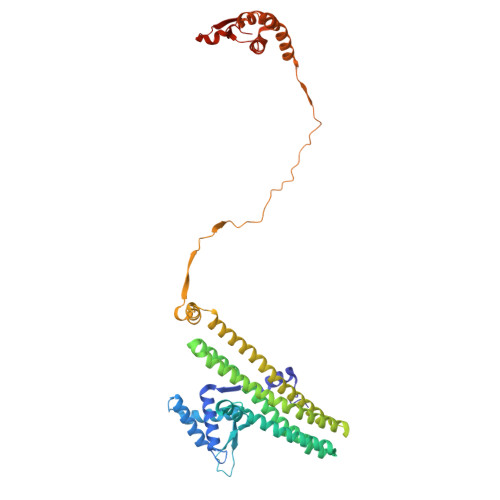
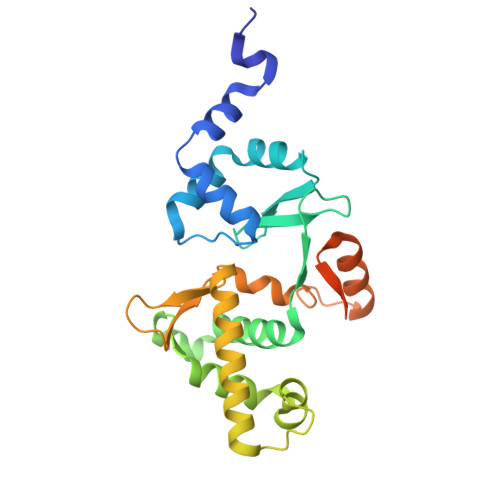
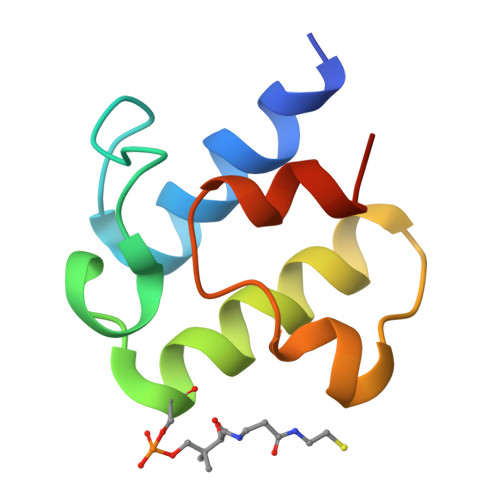
Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic.
Burmann, F., Clifton, B., Koekemoer, S., Wilkinson, O.J., Kimanius, D., Dillingham, M.S., Lowe, J.(2025) Cell 188: 2465
- PubMed: 40168993
- DOI: https://doi.org/10.1016/j.cell.2025.02.032
- Primary Citation of Related Structures:
9GM6, 9GM7, 9GM8, 9GM9, 9GMA, 9GMB, 9GMD - PubMed Abstract:
Ring-like structural maintenance of chromosome (SMC) complexes are crucial for genome organization and operate through mechanisms of DNA entrapment and loop extrusion. Here, we explore the DNA loading process of the bacterial SMC complex MukBEF. Using cryoelectron microscopy (cryo-EM), we demonstrate that ATP binding opens one of MukBEF's three potential DNA entry gates, exposing a DNA capture site that positions DNA at the open neck gate. We discover that the gp5.9 protein of bacteriophage T7 blocks this capture site by DNA mimicry, thereby preventing DNA loading and inactivating MukBEF. We propose a comprehensive and unidirectional loading mechanism in which DNA is first captured at the complex's periphery and then ingested through the DNA entry gate, powered by a single cycle of ATP hydrolysis. These findings illuminate a fundamental aspect of how ubiquitous DNA organizers are primed for genome maintenance and demonstrate how this process can be disrupted by viruses.
- MRC Laboratory of Molecular Biology, Structural Studies, Francis Crick Avenue, Cambridge CB2 0QH, UK; University of Oxford, Department of Biochemistry, South Parks Road, Oxford OX1 3QU, UK. Electronic address: frank.burmann@bioch.ox.ac.uk.
Organizational Affiliation: