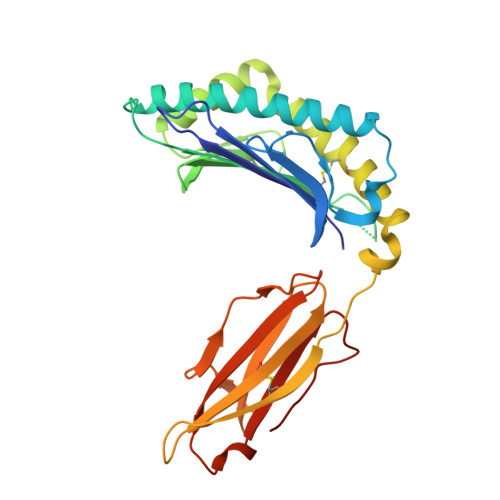
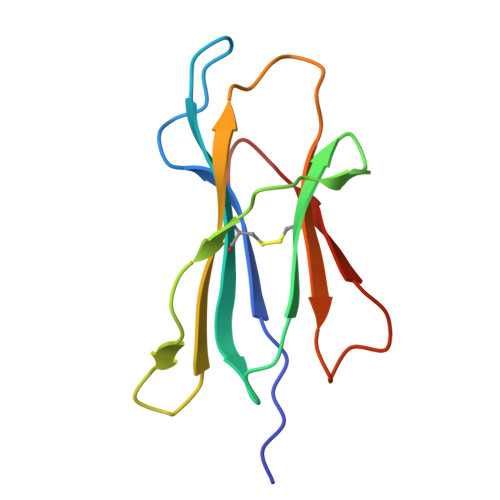
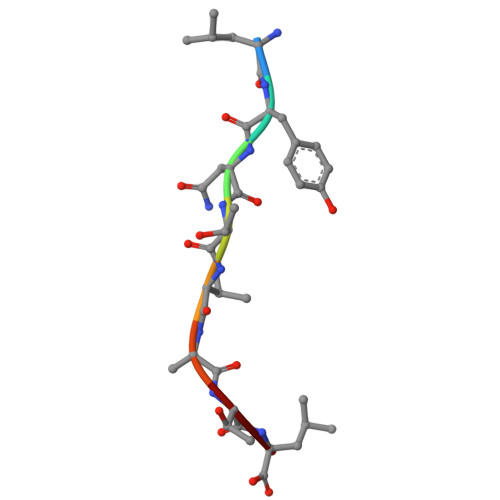
Micropolymorphism outside the peptide-binding groove of human leukocyte antigen (HLA)-C*14 modulates structural stability and shapes immune responses.
Liu, Q., Yang, M., Zhong, P., Wei, Q., Jiao, H., Meng, J., Ding, L., Zhu, X., Wei, P.(2025) Int J Biol Macromol 309: 142772-142772
- PubMed: 40185448
- DOI: https://doi.org/10.1016/j.ijbiomac.2025.142772
- Primary Citation of Related Structures:
9L4G, 9L4H, 9L4I - PubMed Abstract:
Micropolymorphisms in human leukocyte antigen class I (HLA-I) molecules critically influence antigen presentation and immune recognition. Most studies have focused on variations within the peptide-binding groove (PBG), neglecting the potential impact of residues located outside this region. HLA-C*14:02 and HLA-C*14:03 differ only at position 21 (R21 and H21, respectively), which is situated outside the PBG, yet these two allotypes exhibit distinct clinical associations with HIV control in the context of KIR2DL2, an inhibitory killer cell immunoglobulin-like receptor that modulates natural killer (NK) cell activity. Here, we investigated the molecular mechanisms by which the R21H micropolymorphism shapes immune responses. Structural and biochemical analyses revealed that position 21 indirectly regulates the conformation of the B pocket within the PBG, significantly affecting HLA-C*14 stability and altering the composition of its peptide repertoire, while preserving core peptide motifs and recognition by KIR2DL2. Notably, the R21H variation is evolutionarily conserved across various HLA-I molecules and exhibits similar interactions with neighboring residues, suggesting a broadly conserved role in structural stability and immune regulation. These findings suggest that the stability differences between HLA-C*14 allotypes may influence their differential clinical associations, highlighting the previously underappreciated role of micropolymorphisms outside the PBG in modulating immune responses.
- Guangxi Key Laboratory of Special Biomedicine, School of Medicine, Guangxi University, Nanning 530004, China.
Organizational Affiliation: