The structure of FIPV RBD and cat APN complex
Sun, J.Q.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein | 174 | Feline infectious peritonitis virus (strain 79-1146) | Mutation(s): 0 Gene Names: S, 2 | 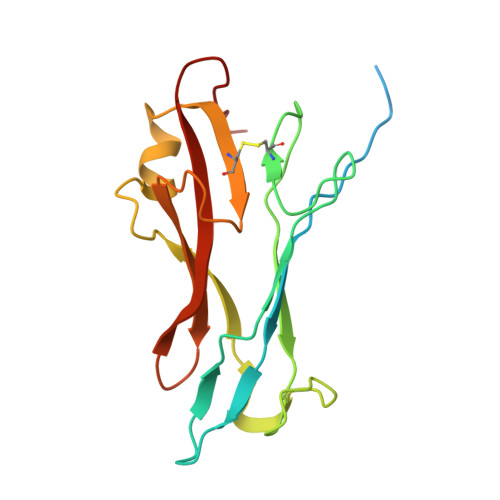 | |
UniProt | |||||
Find proteins for P10033 (Feline coronavirus (strain FIPV WSU-79/1146)) Explore P10033 Go to UniProtKB: P10033 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10033 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Aminopeptidase N | B [auth C] | 932 | Felis catus | Mutation(s): 0 Gene Names: ANPEP, APN EC: 3.4.11.2 | 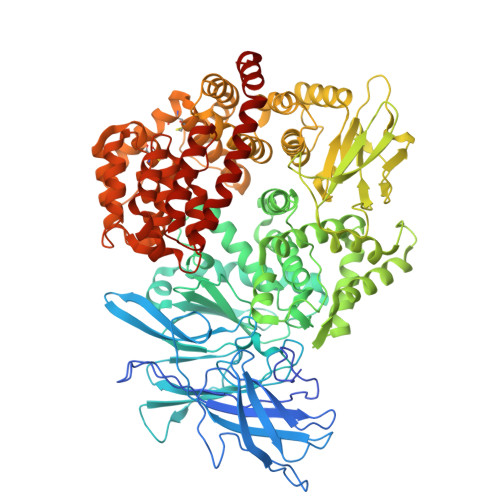 |
UniProt | |||||
Find proteins for P79171 (Felis catus) Explore P79171 Go to UniProtKB: P79171 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P79171 | ||||
Glycosylation | |||||
| Glycosylation Sites: 6 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | F [auth A], G [auth C], H [auth C], I [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32270157 |