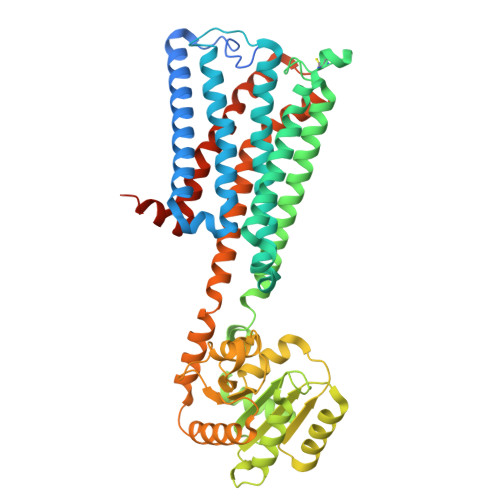
Structural mechanism of CB 1 R binding to peripheral and biased inverse agonists.
Kumari, P., Dvoracsko, S., Enos, M.D., Ramesh, K., Lim, D., Hassan, S.A., Kunos, G., Cinar, R., Iyer, M.R., Rosenbaum, D.M.(2024) Nat Commun 15: 10694-10694
- PubMed: 39695122
- DOI: https://doi.org/10.1038/s41467-024-54206-0
- Primary Citation of Related Structures:
9B9Y, 9B9Z, 9BA0 - PubMed Abstract:
The cannabinoid receptor 1 (CB 1 R) regulates synaptic transmission in the central nervous system, but also has important roles in the peripheral organs controlling cellular metabolism. While earlier generations of brain penetrant CB 1 R antagonists advanced to the clinic for their effective treatment of obesity, such molecules were ultimately shown to exhibit negative effects on central reward pathways that thwarted their further therapeutic development. The peripherally restricted CB 1 R inverse agonists MRI-1867 and MRI-1891 represent a new generation of compounds that retain the metabolic benefits of CB 1 R inhibitors while sparing the negative psychiatric effects. To understand the mechanism of binding and inhibition of CB 1 R by peripherally restricted antagonists, we developed a nanobody/fusion protein strategy for high-resolution cryo-EM structure determination of the GPCR inactive state, and used this method to determine structures of CB 1 R bound to either MRI-1867 or MRI-1891. These structures reveal how these compounds retain high affinity and specificity for CB 1 R's hydrophobic orthosteric site despite incorporating polar functionalities that lead to peripheral restriction. Further, the structure of the MRI-1891 complex along with accompanying molecular dynamics simulations shows how differential engagement with transmembrane helices and the proximal N-terminus can propagate through the receptor to contribute to biased inhibition of β-arrestin signaling.
- Department of Biophysics, The University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: