Search and Browse > Advanced Search
Chemical Similarity Search
Introduction
What is Chemical Similarity Search?
The Chemical Similarity Search option allows you to query the PDB archive using information about small molecules defined in the Chemical Component and BIRD chemical reference dictionaries, such as their molecular formula or chemical descriptors. The search results can be reported either as chemical components that match the query parameters or PDB structures that contain a molecule matching the query criteria (default).
Why run a Chemical Similarity Search?
When you have unique chemical information (e.g., a chemical formula or descriptor) you can use this information to find chemical components (e.g., drugs, inhibitors, modified residues, or building blocks such as amino acids, nucleotides, or sugars), so that it:
- is similar to the formula or descriptor used in the query (perhaps one or two atoms/groups are different)
- is part of a larger molecule (i.e., the specified formula/descriptor is a substructure)
- exactly or very closely matches the formula or descriptor used in the query
The search can also be used to identify PDB structures that include the chemical component(s) which match or are similar to the query. These structures can then be examined to learn about the interactions of the component within the structure.
Documentation
There are a number of different options that can be combined to run a Chemical Similarity Search. These options are being listed here under 3 different sections:
- Query - this will describe the option you have to input your query
- Search - this will describe the types of searches that can be run - from exact match to query, all the way to substructure matches.
- Results - this will describe options available for what you wish to see in the results page.
Query Options
The main ways for initiating a Chemical Similarity Search is either using a Chemical Formula, a descriptor for the molecule, or a 2D chemical drawing.
Chemical Formula
A chemical formula presents the chemical symbols of elements and numbers representing their proportions in the molecule. The order of element symbols in the formula is not important. For example, the input "O1 C12 N4 H28" will match a chemical component with formula "C12 H28 N4 O". Other symbols such as parenthesis, charge indicators may also be included in chemical formulae. Note that a Chemical Formula Search is case-sensitive, so including an uppercase I in the formula "NIC4" will indicate (Nitrogen, Iodine, Carbon4) while a lowercase I indicates "NiC4" (Nickel, Carbon4).
Descriptors
Two main Chemical Descriptors can be used for a chemical component (ligand):
- SMILES (Simplified Molecular Input Line Entry Specification) are chemical notations that allow representation of chemical structures in a way that can be used by the computer. Beyond chemical element symbols, SMILES include a linear notation of molecular structure, including information about bond orders, ring structures, and stereochemistry. Note that SMILES generated by different software may be slightly different. Chemical similarity searches attempt to include most of these representations so SMILES based searches may return more matches than those initiated using InChI descriptors.
- InChI (International Chemical Identifier) - is a standard textual identifier, developed by IUPAC (International Union of Pure and Applied Chemistry) and NIST (National Institute of Standards and Technology), to represent the chemical structure of molecules. This descriptor stores layers of information about the molecules atoms, bond connectivity, stereochemistry, charge etc.
Chemical Drawing
Chemical drawings displays atoms in a ligand, along with their connectivity, bond order, and chirality (as appropriate). You can use the Chemical Sketch tool to draw and edit the 2D structure of a ligand. The tool can automatically convert a chemical drawing into chemical descriptors (SMILES and InChI), and use them to find an exact match or similar molecule in the PDB. Click here to learn more about this tool.
Note: In comparing Chemical Similarity Search results returned from SMILES and InChI descriptor queries, the standard InChI may provide greater specificity. Depending on whether you wish to see fewer or more matches in your results you can use the appropriate descriptor for the search.
Search Options
Using Chemical Formulae
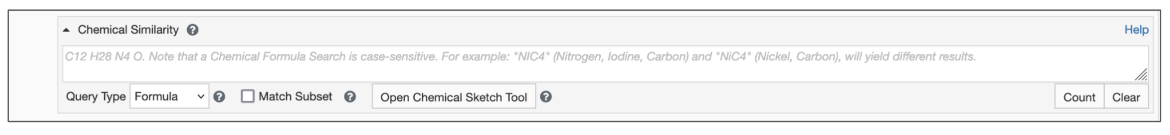
Begin by setting the Query Type to Formula (Figure 1)
- Type the query formula in the text box for Chemical Similarity Search.
- Select the Match Subset options box as appropriate.
|
| Figure 1: Chemical Similarity Search using Chemical Formula |
By default the search will find chemical components whose formula exactly match the query. However, if the Match Subset option is selected, you can select a chemical formula subset which will match any portion of a chemical formula. This option is particularly useful in cases when you are searching for chemical components that include a specific set of elements in a particular ratio. (See example).
Using Chemical Descriptors
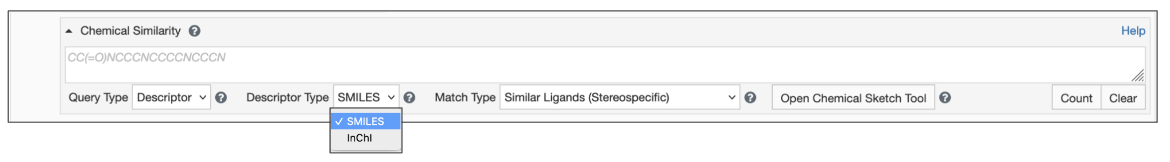
Begin by setting the Query Type to Descriptor (Figure 2)
- Type/paste the chemical descriptor in the text box for Chemical Search.
- Select the appropriate Descriptor Type (SMILES or InChI).
|
| Figure 2: Chemical Similarity Search using Descriptors |
Note: The chemical similarity search descriptor is converted into one of the following a 2D representation for search:
- fingerprints are ordered sets of binary digits (bits) that encode specific physicochemical and/or structural properties of the molecule, such as the presence of common functional groups or ring systems.
- a graph where atoms and bonds in a molecule are mapped onto nodes and edges respectively. Information about atom connectivity, bond order etc. are also coded and used to compare/match different chemical structures.
- Select the Match Type from the options available (see Figure 3):
- Similar Ligands (Quick Screen) - This option uses quick fingerprint matching. The Tanimoto coefficient is used to compute the degree of similarity between a pair of fingerprints. The Tanimoto coefficient has a range from 0 to 1 where higher values indicate greater similarity in structures. Results of Similar Ligands search include molecules with scores exceeding 0.6 for TREE type fingerprints or 0.9 for MACCS type fingerprints. Note that a Tanimoto coefficient of 1 does not indicate a perfect match.
- Similar Ligands (Stereospecific) - in this option the atom type, formal charge, bond order, as well as atom and bond chirality are used as matching criteria. Graph matching is performed on the subset of molecules that satisfy a fingerprint prefilter or screening search. Results will include isomorphic and substructure matches within this screened subset.
- Similar Ligands (including Stereoisomers) in this option the atom type, formal charge, and bond order are used as matching criteria. Graph matching is performed on the subset of molecules that satisfy a fingerprint prefilter or screening search. Results will include isomorphic and substructure matches within this screened subset.
- Substructure (Stereospecific) - in this option graph matching searches perform an exhaustive substructure search where atom type, formal charge, bond order, aromaticity, and atom/bond stereochemistry are used as matching criteria for the search type. Results may include ligands much larger than the query including BIRD molecules where the query molecule is part of the structure.
- Substructure (including Stereoisomers) - in this option graph matching searches perform an exhaustive substructure search where atom type, formal charge, bond order, and aromaticity are used as matching criteria for this search type. Results may include ligands much larger than the query including BIRD molecules where the query molecule is part of the structure.
- Exact match - in this option the atom type, formal charge, aromaticity, bond order, atom/bond stereochemistry, degree, ring membership, and hydrogen count are used as matching criteria for this search type. Results will include chemical components where the query and target graphs match exactly or are very similar. In some cases (especially with SMILES based searches) stereoisomers may also be included in the results.
Result Options
Display Results options before clicking on the query (green magnifying glass) icon.
- Selecting the Structures option will list PDB entries that include the chemical components (ligands or monomers) that match the query
- Selecting the Polymer Entity option will list polymer entities that that have matching monomers in the deposited sequences
- Selecting the Non-polymer Entity option will list non-polymeric small molecules matching the query
- Selecting the Molecular Definitions option will list the chemical components/ligands that match the query
Examples
1. Using Chemical Formula
- C12 H17 N4 O S : will match chemical components with formula "C12 H17 N4 O S" and structures that have those components.
- Ru2 with the Match Subset option selected will match chemical components with a formula containing two rutheniums and structures that have those components.
2. Using Chemical Descriptors
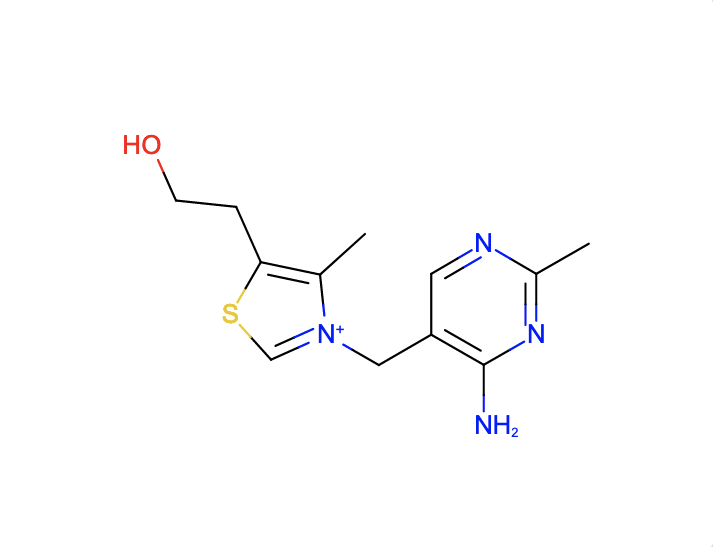
a. Find ligands similar to and with the substructure of the chemical component VIB.
The SMILES search with Cc1c(sc[n+]1Cc2cnc(nc2N)C)CCO and Match Type
- Similar Ligands (Quick screen) will match these chemical components and these structures with those components.
- Similar Ligands (Stereospecific) will match these chemical components and these structures with those components.
- Similar Ligands (including Stereoisomers) will match these chemical components and these structures with those components.
- Substructure (Stereospecific) will match these chemical components and these structures with those components.
- Substructure (including Stereoisomers) will match these chemical components and these structures with those components.
- Exact match will match these chemical components and these structures with those components.
Note that the query descriptor has no chiral atom so there is no difference in the results with the Match Types that are stereospecific and those that include stereoisomers.
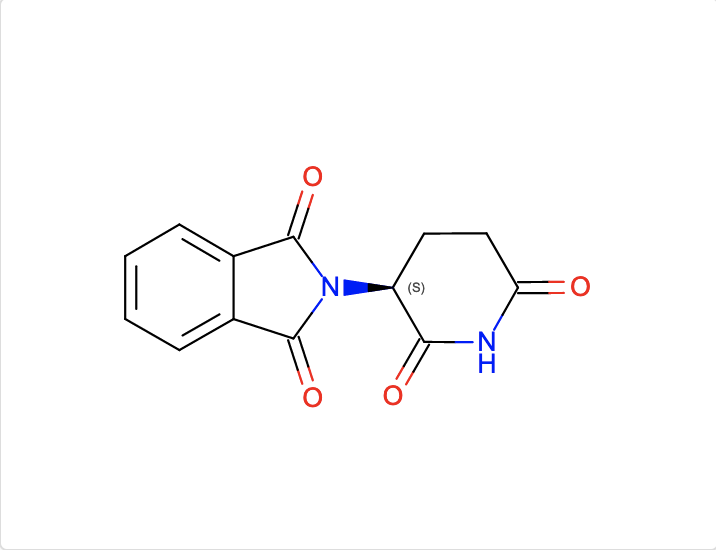
b. Find ligands similar to and with the substructure of the chemical component EF2.
The InChI search with InChI=1S/C13H10N2O4/c16-10-6-5-9(11(17)14-10)15-12(18)7-3-1-2-4-8(7)13(15)19/h1-4,9H,5-6H2,(H,14,16,17)/t9-/m0/s1 and Match Type
- Similar Ligands (Quick screen) will match these chemical components and these structures with those components.
- Similar Ligands (Stereospecific) will match these chemical components and these structures with those components.
- Similar Ligands (including Stereoisomers) will match these chemical components and these structures with those components.
- Substructure (Stereospecific) will match these chemical components and these structures with those components.
- Substructure (including Stereoisomers) will match these chemical components and these structures with those components.
- Exact match will match these chemical components and these structures with those components.
Note that the query descriptor has one chiral atom so the results of the Match Types including and excluding stereoisomers yield different results.