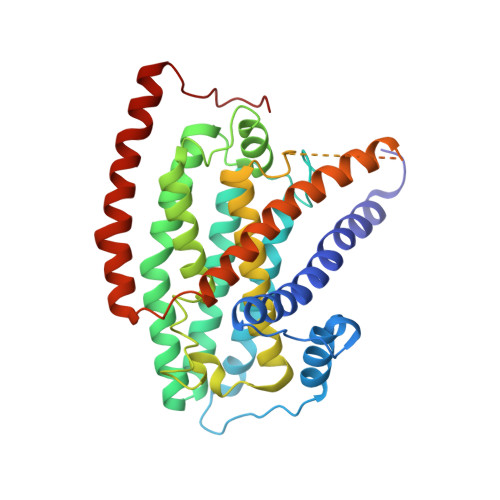
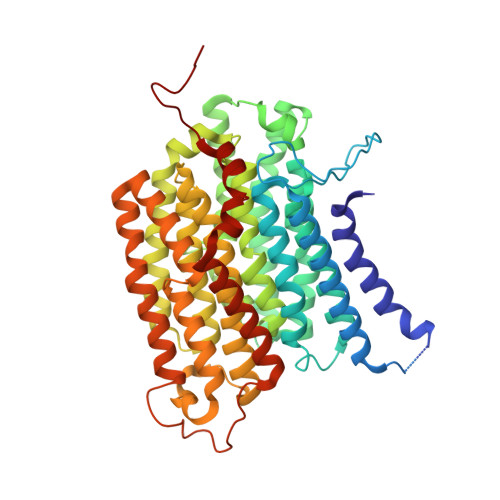
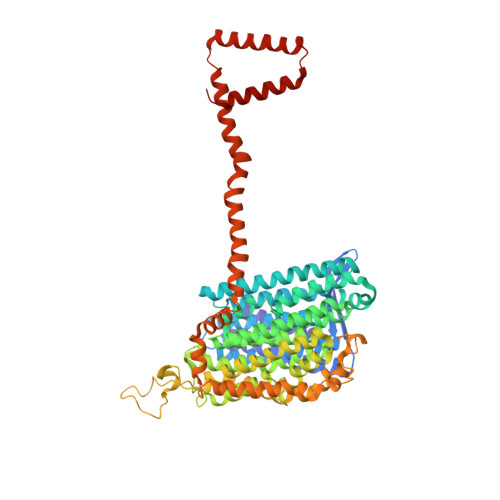
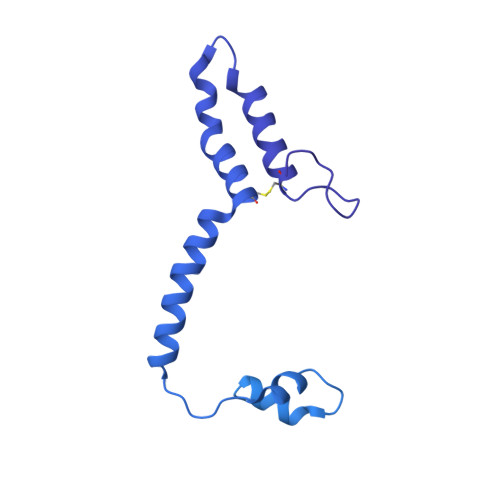
Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Laube, E., Meier-Credo, J., Langer, J.D., Kuhlbrandt, W.(2022) Sci Adv 8: eadc9952-eadc9952
- PubMed: 36427319
- DOI: https://doi.org/10.1126/sciadv.adc9952
- Primary Citation of Related Structures:
7ZM7, 7ZM8, 7ZMB, 7ZME, 7ZMG, 7ZMH - PubMed Abstract:
Mitochondrial complex I is a redox-driven proton pump that generates proton-motive force across the inner mitochondrial membrane, powering oxidative phosphorylation and ATP synthesis in eukaryotes. We report the structure of complex I from the thermophilic fungus Chaetomium thermophilum , determined by cryoEM up to 2.4-Å resolution. We show that the complex undergoes a transition between two conformations, which we refer to as state 1 and state 2. The conformational switch is manifest in a twisting movement of the peripheral arm relative to the membrane arm, but most notably in substantial rearrangements of the Q-binding cavity and the E-channel, resulting in a continuous aqueous passage from the E-channel to subunit ND5 at the far end of the membrane arm. The conformational changes in the complex interior resemble those reported for mammalian complex I, suggesting a highly conserved, universal mechanism of coupling electron transport to proton pumping.
Organizational Affiliation:
Max-Planck-Institute of Biophysics, Frankfurt 60438, Germany.