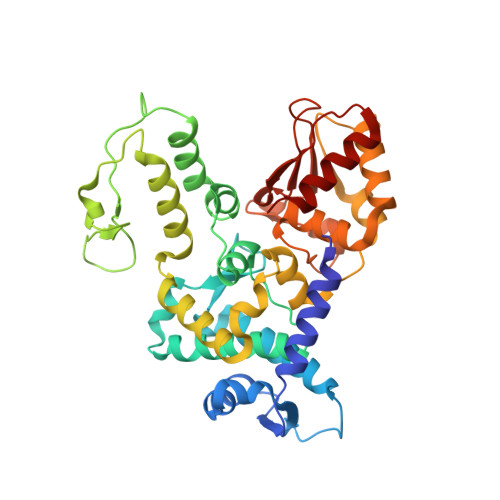
Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase
Verdecia, M.A., Joaziero, C.A.P., Wells, N.J., Ferrer, J.-L., Bowman, M.E., Hunter, T., Noel, J.P.(2003) Mol Cell 11: 249-259
- PubMed: 12535537
- DOI: https://doi.org/10.1016/s1097-2765(02)00774-8
- Primary Citation of Related Structures:
1ND7 - PubMed Abstract:
Ubiquitin ligases (E3) select proteins for ubiquitylation, a modification that directs altered subcellular trafficking and/or degradation of the target protein. HECT domain E3 ligases not only recognize, but also directly catalyze, ligation of ubiquitin to their protein substrates. The crystal structure of the HECT domain of the human ubiquitin ligase WWP1/AIP5 maintains a two-lobed structure like the HECT domain of the human ubiquitin ligase E6AP. While the individual N and C lobes of WWP1 possess very similar folds to those of E6AP, the organization of the two lobes relative to one another is different from E6AP due to a rotation about a polypeptide hinge linking the N and C lobes. Mutational analyses suggest that a range of conformations achieved by rotation about this hinge region is essential for catalytic activity.
Organizational Affiliation:
Structural Biology Laboratory, The Salk Institute for Biological Studies, La Jolla, CA 92037, USA.