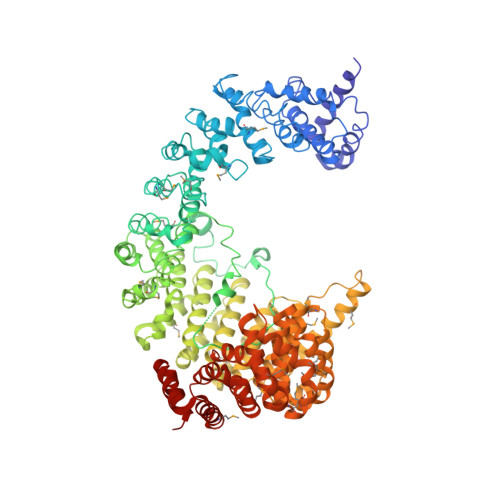
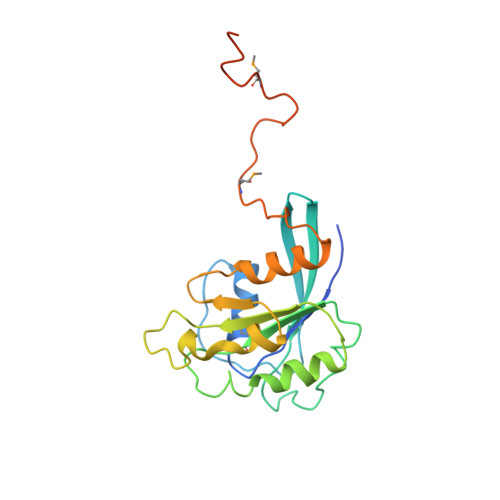
Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp.
Chook, Y.M., Blobel, G.(1999) Nature 399: 230-237
- PubMed: 10353245
- DOI: https://doi.org/10.1038/20375
- Primary Citation of Related Structures:
1QBK - PubMed Abstract:
Transport factors in the karyopherin-beta (also called importin-beta) family mediate the movement of macromolecules in nuclear-cytoplasmic transport pathways. Karyopherin-beta2 (transportin) binds a cognate import substrate and targets it to the nuclear pore complex. In the nucleus, Ran x GTP binds karyopherin-beta2 and dissociates the substrate. Here we present the 3.0 A structure of the karyopherin-beta2-Ran x GppNHp complex where GppNHp is a non-hydrolysable GTP analogue. Karyopherin-beta2 contains eighteen HEAT repeats arranged into two continuous orthogonal arches. Ran is clamped in the amino-terminal arch and substrate-binding activity is mapped to the carboxy-terminal arch. A large loop in HEAT repeat 7 spans both arches. Interactions of the loop with Ran and the C-terminal arch implicate it in GTPase-mediated dissociation of the import-substrate. Ran x GppNHp in the complex shows extensive structural rearrangement, compared to Ran GDP, in regions contacting karyopherin-beta2. This provides a structural basis for the specificity of the karyopherin-beta family for the GTP-bound state of Ran, as well as a rationale for interactions of the karyopherin-Ran complex with the regulatory proteins ranGAP, ranGEF and ranBP1.
- Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, New York, New York 10021, USA.
Organizational Affiliation: