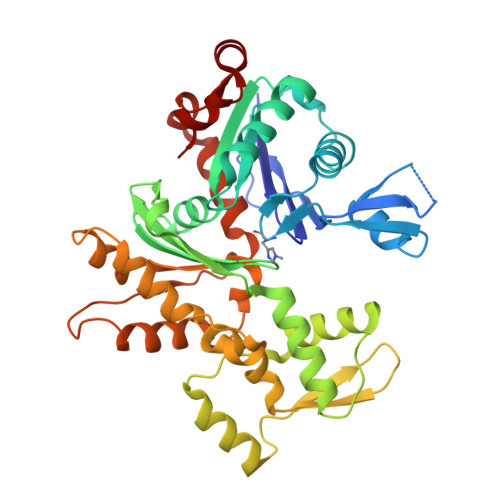
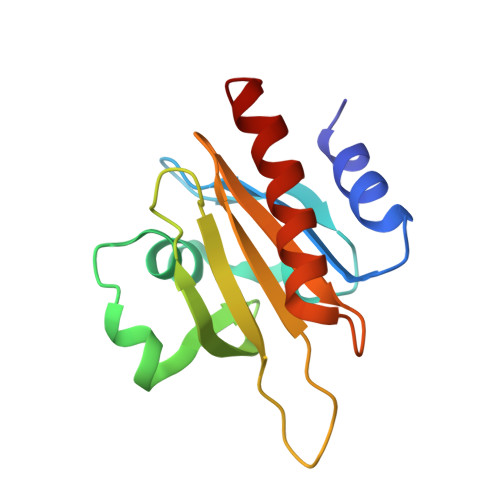
Modulation of actin structure and function by phosphorylation of Tyr-53 and profilin binding.
Baek, K., Liu, X., Ferron, F., Shu, S., Korn, E.D., Dominguez, R.(2008) Proc Natl Acad Sci U S A 105: 11748-11753
- PubMed: 18689676
- DOI: https://doi.org/10.1073/pnas.0805852105
- Primary Citation of Related Structures:
3CHW, 3CI5, 3CIP - PubMed Abstract:
On starvation, Dictyostelium cells aggregate to form multicellular fruiting bodies containing spores that germinate when transferred to nutrient-rich medium. This developmental cycle correlates with the extent of actin phosphorylation at Tyr-53 (pY53-actin), which is low in vegetative cells but high in viable mature spores. Here we describe high-resolution crystal structures of pY53-actin and unphosphorylated actin in complexes with gelsolin segment 1 and profilin. In the structure of pY53-actin, the phosphate group on Tyr-53 makes hydrogen-bonding interactions with residues of the DNase I-binding loop (D-loop) of actin, resulting in a more stable conformation of the D-loop than in the unphosphorylated structures. A more rigidly folded D-loop may explain some of the previously described properties of pY53-actin, including its increased critical concentration for polymerization, reduced rates of nucleation and pointed end elongation, and weak affinity for DNase I. We show here that phosphorylation of Tyr-53 inhibits subtilisin cleavage of the D-loop and reduces the rate of nucleotide exchange on actin. The structure of profilin-Dictyostelium-actin is strikingly similar to previously determined structures of profilin-beta-actin and profilin-alpha-actin. By comparing this representative set of profilin-actin structures with other structures of actin, we highlight the effects of profilin on the actin conformation. In the profilin-actin complexes, subdomains 1 and 3 of actin close around profilin, producing a 4.7 degrees rotation of the two major domains of actin relative to each other. As a result, the nucleotide cleft becomes moderately more open in the profilin-actin complex, probably explaining the stimulation of nucleotide exchange on actin by profilin.
Organizational Affiliation:
Department of Physiology, 3700 Hamilton Walk, University of Pennsylvania School of Medicine, Philadelphia, PA 19104-6085, USA.